DOPA decarboxylase
From Proteopedia
| Line 42: | Line 42: | ||
---- | ---- | ||
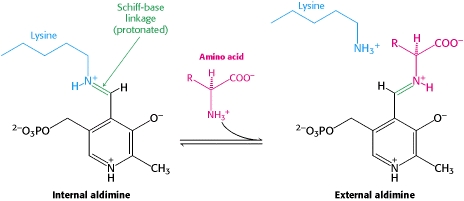
[[image:plp bound.png|thumb|center|400px|'''Schiff base linkage of PLP to Lys303 in the active site''']] | [[image:plp bound.png|thumb|center|400px|'''Schiff base linkage of PLP to Lys303 in the active site''']] | ||
| - | As well, a '''salt bridge''' exists between the carboxyl group of <scene name='DOPA_decarboxylase/Aspartic/1'>Asp271</scene> and the protonated pyridine nitrogen of PLP to further stabilize intermediate. Essentially, a salt bridge combines hydrogen bonding and electrostatic interactions (two common types non-covalent interactions). This interaction serves to provide an electron sink that can stabilize the carbanionic intermediates <ref name="jansonius">PMID:9914259 </ref> . | + | As well, a '''salt bridge''' exists between the carboxyl group of <scene name='DOPA_decarboxylase/Aspartic/1'>Asp271</scene> and the protonated pyridine nitrogen of PLP to further stabilize intermediate. Essentially, a salt bridge combines hydrogen bonding and electrostatic interactions (two common types non-covalent interactions). This interaction serves to provide an electron sink that can stabilize the carbanionic intermediates <ref name="jansonius">PMID:9914259 </ref> . PLP is further anchored to the protein by an extended '''hydrogen bond network''', as shown below. |
| + | ---- | ||
| + | |||
The <scene name='Sandbox/Active_site3/1'>inhibitor</scene> binds to the enzyme by forming a hydrazone linkage. | The <scene name='Sandbox/Active_site3/1'>inhibitor</scene> binds to the enzyme by forming a hydrazone linkage. | ||
Revision as of 20:23, 26 June 2011
Contents |
Introduction
| |||||||||
| 1js3, resolution 2.25Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Activity: | Aromatic-L-amino-acid decarboxylase, with EC number 4.1.1.28 | ||||||||
| Related: | 1js6 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
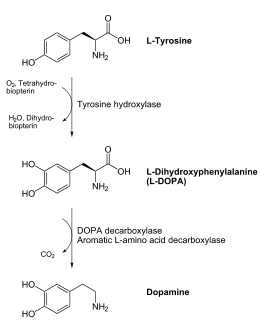
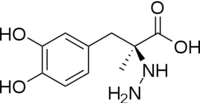
DOPA decarboxylase is responsible for the synthesis of dopamine and serotonin from L-DOPA and L-5-hydroxytryptophan, respectively. Due to its role in neurotransmitter synthesis, DOPA decarboxylase has been implicated in Parkinson's disease, a disease thought to be the result of the degeneration of dopamine-producing cells in the brain. Currently, treatment for the disease is aimed at DOPA decarboxylase inhibition. Since dopamine cannot cross the blood-brain barrier, it cannot be used to directly treat Parkinson's disease. Thus, exogenously administered L-DOPA is the primary treatment for patients suffering from this neurodegenerative disease. Unfortunately, DOPA decarboxylase rapidly converts L-DOPA to dopamine in the blood stream, with only a small percentage reaching the brain. By inhibiting the enzyme, greater amounts of exogenously administered L-DOPA can reach the brain, where it can then be converted to dopamine. [2]
PLP-Dependent Enzymes
Overview
Pyridoxal-5'-phosphate (PLP), the biologically active phosphorylated derivative of vitamin B6, is a versatile and abundant cofactor to a variety of enzymes in all organisms. Almost all PLP-dependet enzymes function in pathways associated with amino compounds, primarily amino acid metabolism. These enzymes carry out many types of reactions, including transaminations, decarboxylations', racemizations, and deaminations (amongst others). PLP is considered to be the most versatile cofactor, thus imparting a wide range of functions on PLP-dependent enzymes. This versatility stems from the fact that PLP can covalently bind the substrate and then act as an electrophilic catalyst. And although these enzymes have wide range of function, there exist only five structural classes: the aspartate amino transferase family, the tryptophan synthase β family, the alanine racemase family, the D-amino acid family, and the glycogen phosphorylase family. [3] [1]The Aspartate Aminotransferase Family
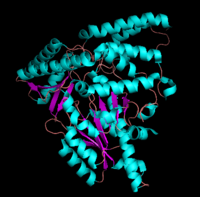
This family of PLP-dependent enzymes is also referred to as fold-type I, with aspartate aminotransferase serving as the prototype. It is the most common structure of the five classes of PLP-dependent enzymes. This fold it is found in a variety of aminotransferases and decarboxylases, amongst them DOPA decarboxylase. PLP-dependent enzymes belonging to this family are catalytically active as homodimers and share a common, well-characterized structure, despite low-sequence identity. Each subunit has a large domain and a small domain. The central feature of the large domain is a seven-stranded β sheet. The small domain has either a three or four-stranded β sheet that is surrounded by α helices on one side. The cofactor PLP is covalently attached to a lysine residue in the large domain and is anchored in a way that allows the aromatic ring of PLP to pack against neighboring β strands. The active site is located in a cleft between the two domains at the interface between the two subunits. Thus, enzymes of fold-type I have residues from both domains and both subunits involved in PLP-binding.
DOPA Decarboxylase
Primary Structure
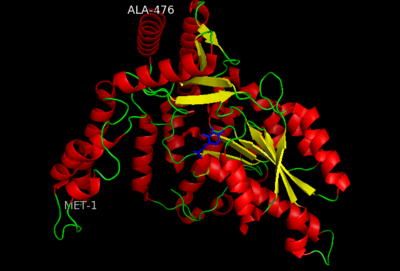
The amino acid sequence of a proteins polypeptide chain is referred to as its primary structure. Each polypeptide chain of DOPA decarboxylase is composed of 486 amino acids that ultimately encode the three-dimensional structure of the protein.
Secondary Structure
The formation of secondary structural elements (like α helices and β sheets) arise in response to the hydrophobic effect and the need to neutralize main-chain polar groups by hydrogen bonding. Each polypeptide chain of DOPA decarboxylase is composed of a seven-stranded mixed , a four-stranded anti-parallel , several , and other, lesser known, secondary structural elements (like loops and the extended strand). Another common secondary structure is the β-turn, or reverse turn. Depicted below is an example of a Type 1 β-turn of DOPA decarboxylase.
This β-turn is comprised of residues Leu-440, Arg-441, Gly-442, and Gln-443. The distance between Cαi and Cαi+3 is 5.1Å, within the acceptable limit of 7Å. As in most β-turns, there is a hydrogen bond between the C=O of Leu-440 and the NH of Gln-443. The phi and psi angles of residues i+1 (Arg-441) and i+2 (Gly-442) are indicated in the diagram.
Tertiary Structure
This level of protein structure refers to the overall three-dimensional shape the polypeptide chain creates. Domains are the fundamental units that generate the tertiary structure, and DOPA decarboxylase is composed of three distinct domains.
The contains the PLP-binding site, and consists of a seven-stranded mixed β sheet that is surrounded by eight α helices, resulting in a typical α/β fold, the most regular and common tertiary structure (recall that α helices and β strands typically alternate in this fold, generating an outer layer of α helices and an inner layer of β sheets). The small is comprised of a four-stranded anti-parallel β sheet that has three α helices packed against the face opposite to the large domain. Although the aforementioned domains exist in all members of this family of PLP-dependent enzymes, including bacterial ornithine decarboxylase (OrnDC) and dialkylglycine decarboxylase (DGD), the is unique to DOPA decarboxylase, and is a representative case of domain swapping. This domain is composed of two parallel helices linked by an extended strand, which essentially lies like a flap over the second subunit. As well, residues from the N-terminal domain and the small domain form a short
Quaternary Structure
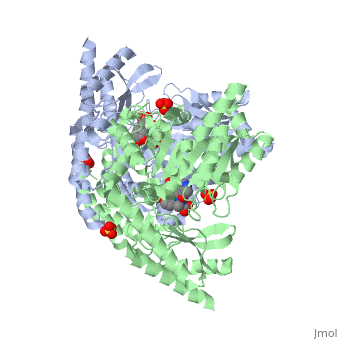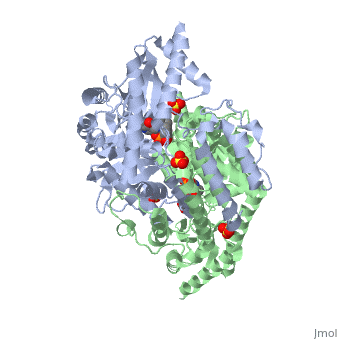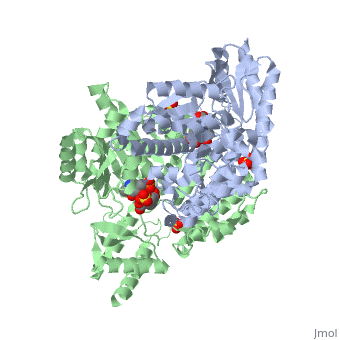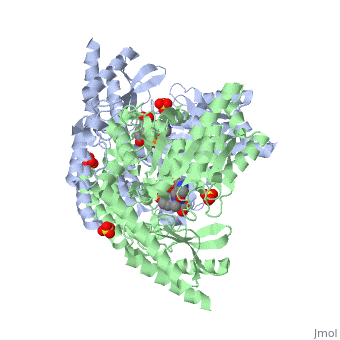
The level of protein structure exists solely in multisubunit complexes. DOPA decarboxylase is a homodimeric enzyme with the active site located near the monomer-monomer interface, thus highlighting the importance of this level of protein structure to the enzymes function. Furthermore, since the N-terminal domain of one monomer packs on top of the other monomer, resulting in an extended dimer interface, this level of tertiary structure is most likely stable only in the dimeric form of the enzyme.
Function
The Active Site
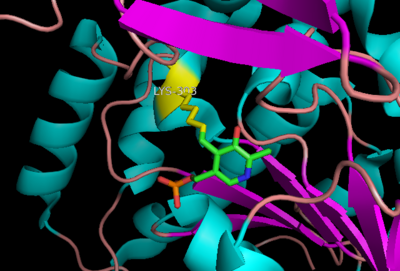
The active site of DOPA decarboxylase is located in a cleft at the between the two subunits of the dimer, like all PLP-dependent enzymes of the aspartate aminotransferase family. Since it is at the interface, residues from both domains and both subunits are involved in cofactor binding, although the active site is composed of residues mainly from one monomer. The is composed of several key residues. serves to bind PLP via a Schiff base linkage.
As well, a salt bridge exists between the carboxyl group of and the protonated pyridine nitrogen of PLP to further stabilize intermediate. Essentially, a salt bridge combines hydrogen bonding and electrostatic interactions (two common types non-covalent interactions). This interaction serves to provide an electron sink that can stabilize the carbanionic intermediates [4] . PLP is further anchored to the protein by an extended hydrogen bond network, as shown below.
The binds to the enzyme by forming a hydrazone linkage.
Substrate Binding
Classification
SCOP
DOPA decarboxylase is classified in the following manner using SCOP:
- Class: alpha and beta proteins (α/β)
- Fold: PLP-dependent transferase-like
- Superfamily: PLP-dependent transferases
- Family: Pyridoxal-dependent decarboxylase
- Domain: DOPA decarboxylase
CATH
DOPA decarboxylase is classified in the following manner using CATH:
- large domain
- Class: alpha beta
- Architecture: 3-layer sandwich
- Topology: Aspartate aminotransferase
- small domain
- Class: alpha beta
- Architecture: alpha-beta complex
- Topology: Aspartate aminotransferase
- N-terminal domain
- Class: mainly alpha
- Architecture: up-down bundle
- Topology: dopa decarboxylase
3D structures of DOPA decarboxylase
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez