This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:JBIC:11
From Proteopedia
(Difference between revisions)

| Line 4: | Line 4: | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| - | |||
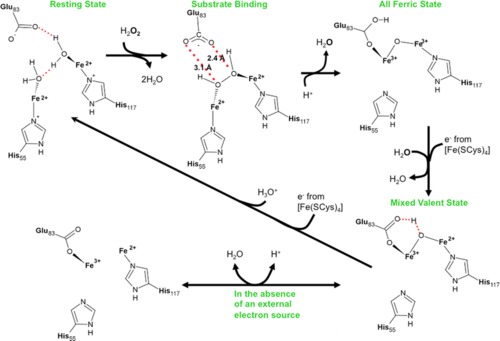
| - | [[Image:Xc.PNG|left|500px|thumb|Reaction scheme for rubrerythrins.]] | ||
Oxidative damage is a major problem for all organisms and not surprisingly a number of protection systems exist in cells across all kingdoms of life. For organisms that respire using anaerobic processes oxidative damage can be an even more pressing problem as these cells may be destroyed by the simple presence of molecular oxygen. It is not that surprising that strict anaerobes have developed a system for the rapid reduction and removal of molecular oxygen in two steps. The first involves reduction of oxygen to peroxide followed by peroxide reduction to water. The terminal enzyme in this systems, rubrerythrin, has been well characterized in a number of bacteria that live at ambient temperatures. In this work, we report the first time course that monitors peroxide reduction by a rubrerythrin isolated from an organism that lives at an optimal temperature of one-hundred degrees centigrade. The slow turnover of the enzyme at room temperature has allowed us to trap intermediates and track the atomic details of this important enzyme reaction. | Oxidative damage is a major problem for all organisms and not surprisingly a number of protection systems exist in cells across all kingdoms of life. For organisms that respire using anaerobic processes oxidative damage can be an even more pressing problem as these cells may be destroyed by the simple presence of molecular oxygen. It is not that surprising that strict anaerobes have developed a system for the rapid reduction and removal of molecular oxygen in two steps. The first involves reduction of oxygen to peroxide followed by peroxide reduction to water. The terminal enzyme in this systems, rubrerythrin, has been well characterized in a number of bacteria that live at ambient temperatures. In this work, we report the first time course that monitors peroxide reduction by a rubrerythrin isolated from an organism that lives at an optimal temperature of one-hundred degrees centigrade. The slow turnover of the enzyme at room temperature has allowed us to trap intermediates and track the atomic details of this important enzyme reaction. | ||
| Line 11: | Line 9: | ||
state results in a <scene name='Journal:JBIC:11/Cv/10'>significant distortion of an alpha-helix</scene>. The <scene name='Journal:JBIC:11/Cv/11'>movement of backbone carbon atoms is over 1.7 A</scene> in some cases. <scene name='Journal:JBIC:11/Exp/1'>10-s exposure of rubrerythrin crystals to peroxide</scene> reveals <scene name='Journal:JBIC:11/Exp/2'>small srtuctural changes</scene> ([[3pza]]). <scene name='Journal:JBIC:11/Exp/3'>15-s explosure</scene> also reveals <scene name='Journal:JBIC:11/Exp/4'>small srtuctural changes</scene> ([[3qvd]]). Both these exposures identified previously unobserved intermediates in the reaction cycle. | state results in a <scene name='Journal:JBIC:11/Cv/10'>significant distortion of an alpha-helix</scene>. The <scene name='Journal:JBIC:11/Cv/11'>movement of backbone carbon atoms is over 1.7 A</scene> in some cases. <scene name='Journal:JBIC:11/Exp/1'>10-s exposure of rubrerythrin crystals to peroxide</scene> reveals <scene name='Journal:JBIC:11/Exp/2'>small srtuctural changes</scene> ([[3pza]]). <scene name='Journal:JBIC:11/Exp/3'>15-s explosure</scene> also reveals <scene name='Journal:JBIC:11/Exp/4'>small srtuctural changes</scene> ([[3qvd]]). Both these exposures identified previously unobserved intermediates in the reaction cycle. | ||
| + | [[Image:Xc.PNG|left|500px|thumb|Reaction scheme for rubrerythrins.]] | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Revision as of 10:41, 5 July 2011
| |||||||||||
- ↑ Dillard BD, Demick JM, Adams MW, Lanzilotta WN. A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus. J Biol Inorg Chem. 2011 Jun 7. PMID:21647777 doi:10.1007/s00775-011-0795-6
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.