Journal:Cell:1
From Proteopedia
(Difference between revisions)

| Line 10: | Line 10: | ||
===Interactions Between IFNAR & IFN=== | ===Interactions Between IFNAR & IFN=== | ||
====IFNAR2-IFN interaction==== | ====IFNAR2-IFN interaction==== | ||
| - | [[Image: | + | [[Image:IFNa_IFNAR2_interaction_map.png|300px||right|]] |
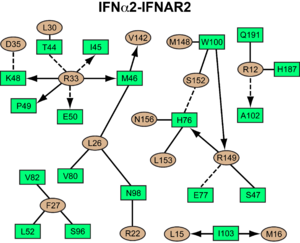
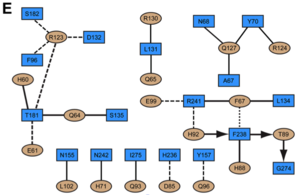
<scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33IFN appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2 (Figure 3B and D). It forms an <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_cartoon/2'>extensive hydrogen-bonding network</scene> with the main chain carbonyl oxygen atoms of <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_non_cartoon/3'>Ile45, Lys48, Glu50 and the side chain of Thr44</scene>. This residue is present in IFNα, IFNω, IFNβ and IFNε. Two hydrophobic interaction clusters are present in the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interact_hydro_full/1'>IFNa-IFNAR2</scene> interface: <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop1/3'>the first one</scene> is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; Ala19IFN and Met148IFN line this cluster; <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop2/1'>the second one</scene> comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor (Figure 3D). Replacing <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30/1'>Leu30IFN with alanine</scene> reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30_nono/4'>not engaged in any intimate contacts with IFNAR2 residues</scene>. One reason for its importance might be a <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_arg_stabilized/1'>stabilizing effect on the position of Arg33IFN</scene>. Furthermore, hydrogen bonds in the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_preh/3'>IFNa2-IFNAR2 interface</scene> are observed between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_h76_hbonds/3'>Ser152IFN and His76R2</scene>, and <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_h76_hbonds2/1'>Arg22IFN and Asn98R2</scene> (Figure 3D). [[Image:Workbench 3E.png|300px|left|]] Most of the residues involved in the IFNα2-IFNAR2 interaction are also found in the IFNw1-IFNAR2 interface of the IFNω1 ternary complex (Figure 3E). A significant difference in the IFNAR2 interface between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_dif/5'>IFNa2</scene> and IFNω is related to <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_salt/1'>Arg149 in IFNa2</scene>, which is replaced with Lys152 in <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw</scene>. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_interface/3'>IFNw1-IFNAR2 interface</scene>, this residue forms an <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_salt/1'>intramolecular salt bridge</scene> with Glu147IFN, but <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_no_interact/1'>does not contact Glu77 of the receptor</scene>. The IFNω1-IFNAR2 interface buries 1820 Å2 of surface area. | <scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33IFN appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2 (Figure 3B and D). It forms an <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_cartoon/2'>extensive hydrogen-bonding network</scene> with the main chain carbonyl oxygen atoms of <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_non_cartoon/3'>Ile45, Lys48, Glu50 and the side chain of Thr44</scene>. This residue is present in IFNα, IFNω, IFNβ and IFNε. Two hydrophobic interaction clusters are present in the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interact_hydro_full/1'>IFNa-IFNAR2</scene> interface: <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop1/3'>the first one</scene> is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; Ala19IFN and Met148IFN line this cluster; <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop2/1'>the second one</scene> comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor (Figure 3D). Replacing <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30/1'>Leu30IFN with alanine</scene> reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30_nono/4'>not engaged in any intimate contacts with IFNAR2 residues</scene>. One reason for its importance might be a <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_arg_stabilized/1'>stabilizing effect on the position of Arg33IFN</scene>. Furthermore, hydrogen bonds in the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_preh/3'>IFNa2-IFNAR2 interface</scene> are observed between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_h76_hbonds/3'>Ser152IFN and His76R2</scene>, and <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_h76_hbonds2/1'>Arg22IFN and Asn98R2</scene> (Figure 3D). [[Image:Workbench 3E.png|300px|left|]] Most of the residues involved in the IFNα2-IFNAR2 interaction are also found in the IFNw1-IFNAR2 interface of the IFNω1 ternary complex (Figure 3E). A significant difference in the IFNAR2 interface between <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_dif/5'>IFNa2</scene> and IFNω is related to <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction_salt/1'>Arg149 in IFNa2</scene>, which is replaced with Lys152 in <scene name='User:David_Canner/Workbench2/Ifnw_ifnar21_structure/3'>IFNw</scene>. In the <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_interface/3'>IFNw1-IFNAR2 interface</scene>, this residue forms an <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_salt/1'>intramolecular salt bridge</scene> with Glu147IFN, but <scene name='User:David_Canner/Workbench2/Ifnw_ifnar2_no_interact/1'>does not contact Glu77 of the receptor</scene>. The IFNω1-IFNAR2 interface buries 1820 Å2 of surface area. | ||
Revision as of 05:02, 25 July 2011
| |||||||||||
- ↑ no reference
Proteopedia Page Contributors and Editors (what is this?)
Christoph Thomas, Jaime Prilusky, Joel L. Sussman, Michal Harel, Alexander Berchansky
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.