Journal:Cell:1
From Proteopedia
(Difference between revisions)

| Line 6: | Line 6: | ||
IFNs were the first cytokines discovered more than half a century ago as agents that interfere with viral infection. Since then, IFNs have been established as pleiotropic, multifunctional proteins in the early immune response, exhibiting pronounced antiproliferative effects on cells, in addition to their strong immunomodulatory and antiviral activity. Due to their potency and diverse biological activities, IFNs are used for the treatment of several human diseases, including hepatitis C, multiple sclerosis and certain types of cancer. | IFNs were the first cytokines discovered more than half a century ago as agents that interfere with viral infection. Since then, IFNs have been established as pleiotropic, multifunctional proteins in the early immune response, exhibiting pronounced antiproliferative effects on cells, in addition to their strong immunomodulatory and antiviral activity. Due to their potency and diverse biological activities, IFNs are used for the treatment of several human diseases, including hepatitis C, multiple sclerosis and certain types of cancer. | ||
| - | All <scene name='User:David_Canner/Workbench/Opening_ifna/2'>type I IFNs</scene> initiate signaling by binding to the same receptor composed of two subunits called <scene name='User:David_Canner/Workbench/Opening_ifnar1/3'>IFNAR1</scene> and <scene name='User:David_Canner/Workbench/Opening_ifnar2/2'>IFNAR2</scene>. The intracellular domains (ICDs) of IFNAR1 and IFNAR2 are associated with the Janus kinases (Jaks) Tyk2 and Jak1, respectively. Upon ligand binding by the IFNAR chains and formation of the signaling complex, these tyrosine kinases trans-phosphorylate and thereby activate each other. Subsequently, the activated Jaks phosphorylate STAT1, STAT2 and STAT3, which translocate into the nucleus and activate the transcription of hundreds of IFN-stimulated genes. To gain insight into how type I IFNs engage their receptor chains, how the receptor system is able to recognize the large number of different ligands, and how different IFN ligands can evoke different physiological activities, we determined the crystal structures of unliganded <scene name='User:David_Canner/Workbench/Opening_ifnar1_alone/2'>IFNAR1 (SD1 through SD3)</scene>, the binary complex <scene name='User:David_Canner/Workbench/Opening_ifnar2_binary/1'>between | + | All <scene name='User:David_Canner/Workbench/Opening_ifna/2'>type I IFNs</scene> initiate signaling by binding to the same receptor composed of two subunits called <scene name='User:David_Canner/Workbench/Opening_ifnar1/3'>IFNAR1</scene> and <scene name='User:David_Canner/Workbench/Opening_ifnar2/2'>IFNAR2</scene>. The intracellular domains (ICDs) of IFNAR1 and IFNAR2 are associated with the Janus kinases (Jaks) Tyk2 and Jak1, respectively. Upon ligand binding by the IFNAR chains and formation of the signaling complex, these tyrosine kinases trans-phosphorylate and thereby activate each other. Subsequently, the activated Jaks phosphorylate STAT1, STAT2 and STAT3, which translocate into the nucleus and activate the transcription of hundreds of IFN-stimulated genes. To gain insight into how type I IFNs engage their receptor chains, how the receptor system is able to recognize the large number of different ligands, and how different IFN ligands can evoke different physiological activities, we determined the crystal structures of unliganded <scene name='User:David_Canner/Workbench/Opening_ifnar1_alone/2'>IFNAR1 (SD1 through SD3)</scene>, the binary complex <scene name='User:David_Canner/Workbench/Opening_ifnar2_binary/1'>between IFNa2 and IFNAR2</scene>, and the ternary ligand-receptor complexes of <scene name='User:David_Canner/Workbench/Opening_ternary_alpha/2'>IFNa2</scene> and <scene name='User:David_Canner/Workbench/Opening_ternary_gamma/3'>IFNw</scene> binding both receptor chains. A final theoretical ternary structure including <scene name='User:David_Canner/Workbench/Opening_sd4_ternary/1'>the membrane-proximal sub-domain (SD4) of IFNAR1</scene> was also created. These structures, in conjunction with biochemical and cellular experiments, reveal that the type I IFN receptor uses a mode of ligand interaction that is unique among cytokine receptors, but conserved between different IFNs. |
===Interactions Between IFNAR & IFN=== | ===Interactions Between IFNAR & IFN=== | ||
====IFNAR2-IFN interaction==== | ====IFNAR2-IFN interaction==== | ||
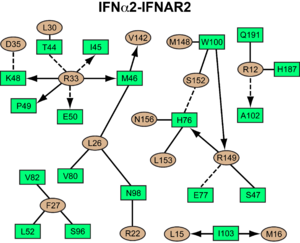
[[Image:IFNa_IFNAR2_interaction_map.png|300px||right|]] | [[Image:IFNa_IFNAR2_interaction_map.png|300px||right|]] | ||
| - | <scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33(IFN) appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2. It forms an <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_cartoon/2'>extensive hydrogen-bonding network</scene> with the main chain carbonyl oxygen atoms of <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_non_cartoon/3'>Ile45(IFNAR2) and Glu50(IFNAR2) and the side chain of Thr44(IFNAR2)</scene>. This residue is present in | + | <scene name='User:David_Canner/Workbench/Opening_ifna/2'>Interferon</scene> interacts primarily with the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interaction/1'>D1 domain of IFNAR2</scene>. Arg33(IFN) appears to be the <scene name='User:David_Canner/Workbench2/Ifn_arg_33/1'>single most important residue</scene> for the interaction of the IFN ligand with IFNAR2. It forms an <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_cartoon/2'>extensive hydrogen-bonding network</scene> with the main chain carbonyl oxygen atoms of <scene name='User:David_Canner/Workbench2/Ifn_h_bonds_non_cartoon/3'>Ile45(IFNAR2) and Glu50(IFNAR2) and the side chain of Thr44(IFNAR2)</scene>. This residue is present in IFNa, IFNw, IFNb and IFNe. Two hydrophobic interaction clusters are present in the <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_interact_hydro_full/1'>IFNa-IFNAR2</scene> interface: <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop1/3'>the first one</scene> is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_hydrop2/1'>the second one</scene> comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor. Replacing <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30/1'>Leu30(IFN) with alanine</scene> reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_leu_30_nono/4'>not engaged in any intimate contacts with IFNAR2 residues</scene>. One reason for its importance might be a <scene name='User:David_Canner/Workbench2/Ifn_ifnar2_arg_stabilized/1'>stabilizing effect on the position of Arg33(IFN)</scene>. |
Most of the residues involved in the IFNα2-IFNAR2 interaction are also found in the IFNω-IFNAR2 interface of the IFNω ternary complex. | Most of the residues involved in the IFNα2-IFNAR2 interaction are also found in the IFNω-IFNAR2 interface of the IFNω ternary complex. | ||
[[Image:IFNw_IFNAR2_interaction_map.png|300px|left|]] | [[Image:IFNw_IFNAR2_interaction_map.png|300px|left|]] | ||
Revision as of 08:24, 25 July 2011
| |||||||||||
- ↑ no reference
Proteopedia Page Contributors and Editors (what is this?)
Christoph Thomas, Jaime Prilusky, Joel L. Sussman, Michal Harel, Alexander Berchansky
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.