Ouabain
From Proteopedia
| Line 8: | Line 8: | ||
==Ouabain Structure and Binding== | ==Ouabain Structure and Binding== | ||
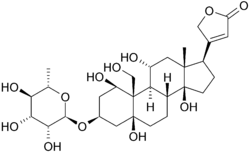
| - | Below is the structure of oubain in two dimensions. The molecule consists of a sugar bound to a modified cholesterol by a glycosidic linkage (hence ''glycoside''). The hydroxyl groups surrounding much of the molecule, along with the esters at either end, contribute to its binding to the membrane bound sodium-potassium pump. <scene name='Sandbox_60/Ouabain_3d/1'>Ouabain</scene> can also be seen in three dimensions. With the molecular geometry and stereochemistry displayed in this way, one can see more clearly the distribution of polar carbon-oxygen and non-polar carbon-carbon bonds in the space surrounding the molecule. This makes visualizing the binding of the inhibitor much easier. | + | [[Image:Ouabain.png|250px|left|upright=1.5]] Below is the structure of oubain in two dimensions. The molecule consists of a sugar bound to a modified cholesterol by a glycosidic linkage (hence ''glycoside''). The hydroxyl groups surrounding much of the molecule, along with the esters at either end, contribute to its binding to the membrane bound sodium-potassium pump. <scene name='Sandbox_60/Ouabain_3d/1'>Ouabain</scene> can also be seen in three dimensions. With the molecular geometry and stereochemistry displayed in this way, one can see more clearly the distribution of polar carbon-oxygen and non-polar carbon-carbon bonds in the space surrounding the molecule. This makes visualizing the binding of the inhibitor much easier. Ouabain is <scene name='Sandbox_60/Drug_in_complex/1'>bound</scene> to the protein along the inside of an alpha-helix bundle. <scene name='Sandbox_60/Drug_in_complex_np/1'>Non-polar</scene> components of residues help somewhat in coordinating ouabain through Van der Waals forces, but <scene name='Sandbox_60/Drug_in_complex_p/1'>polar</scene> residues, glutamine, aspartic acid, and threonine, along with the amide bond of an alanine, surround the hydroxyl and carbonyl groups of the ligand, forming hydrogen bonds of 2 to 4 angstroms in length. This not only holds the drug in place, but prevents conformational change necessary for the function of the protein. |
==Cardiac Muscle and Ion Pump Inhibition== | ==Cardiac Muscle and Ion Pump Inhibition== | ||
Revision as of 20:43, 15 August 2011
| |||||||||||
Sources
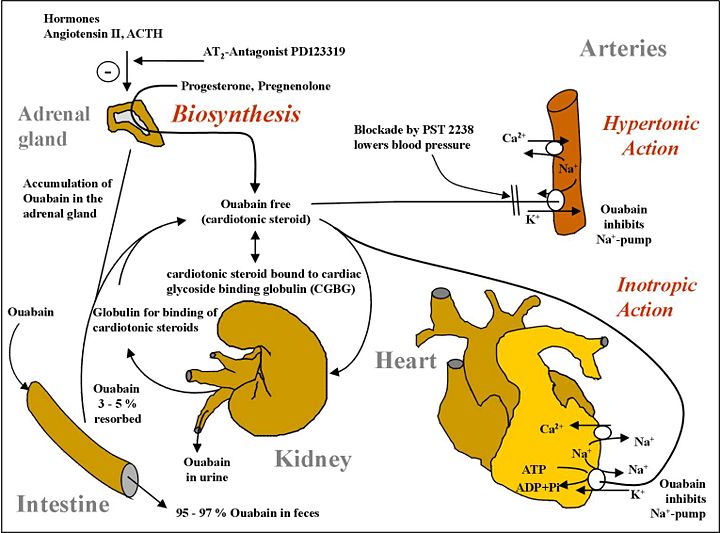
It was recently discovered that ouabain, long thought to be exclusively a plant product, is actually synthesized by animals, and secreted from the adrenal cortex to regulate body osmosis and cellular concentrations of sodium. The image below represents its biosynthesis and metabolism in humans.
Though there are currently synthetic schemes for the production of Ouabain, the compound is usually extracted from the plant sources Strophanthus gratus (left) and Acokanthera schimperi. Somalia is both the native habitat of these plants and the etymological origin of the name ouabain. Somalian tribes have historically used ouabain poisoned arrows for hunting. These arrows are capable of killing a hippopotamus, likely due to cardiac arrest.