User:R. Jeremy Johnson/RNaseA
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
==='''Active Site Structure'''=== | ==='''Active Site Structure'''=== | ||
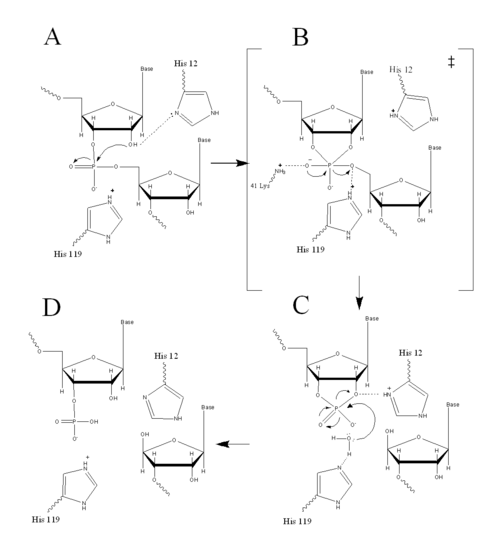
| - | [[Image:MechIII.png| | + | [[Image:MechIII.png|500px|left|thumb|Figure II: RNase A Catalysis. (A) Initial attack of 2'hydroxyl stabilized by His12. (B) Pentavalent phosphorous intermediate. (C) 2'3' cyclic intermediate degradation. (D) Finished products: Two distinctive nucleotide sequences. Figure generated via ''Chemdraw'']] RNase A uses acid/base catlysis to speed up RNA hydrolysis. This occurs in the <scene name='Sandbox_Reserved_193/Active_site_a/1'>active site</scene> which is found in the cleft of RNase A and is the location of the chemical change in bound substrates. Subsites lining the active site cleft are important to the binding of single stranded RNA. Large quantities of positively charged residues, such as <scene name='Sandbox_Reserved_193/Lys7_arg10_arg39_lys41_lys66/1'>Lys7, Arg10, Arg39, and Lys41, and Lys66</scene>, recognize the negative charge on the phosphate back bone of the <scene name='Sandbox_Reserved_194/Positive_amino_acids_substrate/1'>RNA strand</scene> <ref name="Wlodrawer" />. |
The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. <scene name='Sandbox_Reserved_193/Thr45_a/1'>Threonine 45</scene>, located next to the active site, will hydrogen bond to pyrimidine bases, but sterically hinder the binding of a purine on the 5' strand of OH. Thr45 significantly decreases the rate of hydrolysis of polymeric purine strands, such as poly A, by a thousand fold, as compared to polymeric pyrimidine strands.<ref name = 'Wlodrawer'>PMID:3401445</ref> | The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. <scene name='Sandbox_Reserved_193/Thr45_a/1'>Threonine 45</scene>, located next to the active site, will hydrogen bond to pyrimidine bases, but sterically hinder the binding of a purine on the 5' strand of OH. Thr45 significantly decreases the rate of hydrolysis of polymeric purine strands, such as poly A, by a thousand fold, as compared to polymeric pyrimidine strands.<ref name = 'Wlodrawer'>PMID:3401445</ref> | ||
| - | Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold <ref name="Wlodrawer" />. | + | Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold <ref name="Wlodrawer" />. |
| + | |||
| + | ==='''Acid Base Catalysis'''=== | ||
| + | In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. RNase A also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (p''K''a=6); therefore, histidine can both accept and donate protons at physiological pH. | ||
| + | |||
| + | Acid/base catalysis by an enzyme is dependent on the pH of the environment and the p''K''a's of their residues. The pKa value will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the p''K''a. This ability to alter the pKa of certain residues such as histidines, increases the diversity of reactions that an enzyme can perform <ref name = 'Lehninger'>'Lehninger A., Nelson D.N, & Cox M.M. (2008) Lehninger Principles of Biochemistry. W. H. Freeman, fifth edition.' </ref>. | ||
==='''Acid Base Catalysis by RNase A'''=== | ==='''Acid Base Catalysis by RNase A'''=== | ||
| - | RNase A catalyzes the cleavage of the Phosphodiester bonds in two steps: the formation of the pentavalent phosphate transition state and subsequent degradation of the 2’3’ cyclic phosphate intermediate. An important part of the reaction is the ability of histidine (His 12 and His119) to both accept and donate electrons, allowing these histidine to be an acid or a base, making the reaction pH dependent <ref name="Raines" />. | + | RNase A catalyzes the cleavage of the Phosphodiester bonds in two steps: the formation of the pentavalent phosphate transition state and subsequent degradation of the 2’3’ cyclic phosphate intermediate using three main catalytic residues (<scene name='Sandbox_Reserved_193/His12_lys41_his119/1'>His12, Lys41, and His119</scene> and <scene name='Sandbox_Reserved_194/His12_lys41_his119/1'>His12, Lys41, and His119 with substrate present</scene>). An important part of the reaction is the ability of histidine (His 12 and His119) to both accept and donate electrons, allowing these histidine to be an acid or a base, making the reaction pH dependent <ref name="Raines" />. |
RNA hydrolysis begins when <scene name='Sandbox_Reserved_193/His12a_a/1'>His12</scene> abstracts a proton from the 2’ OH group on RNA; thus, assisting in the nucleophilic attack of the 2’ oxygen on the electrophilic phosphorus atom. A transition state is then formed, having a pentavalent phosphate, which is stabilized by the positively charged amino group of <scene name='Sandbox_Reserved_193/Lys41a_a/1'>Lys41</scene> and the main chain amide nitrogen of Phe120. <scene name='Sandbox_Reserved_193/His119a_a/1'>His119</scene> then protonates the 5' oxygen on the ribose ring and the transition state falls to form a 2’3’cyclic phosphate intermediate <ref name="Raines" />. | RNA hydrolysis begins when <scene name='Sandbox_Reserved_193/His12a_a/1'>His12</scene> abstracts a proton from the 2’ OH group on RNA; thus, assisting in the nucleophilic attack of the 2’ oxygen on the electrophilic phosphorus atom. A transition state is then formed, having a pentavalent phosphate, which is stabilized by the positively charged amino group of <scene name='Sandbox_Reserved_193/Lys41a_a/1'>Lys41</scene> and the main chain amide nitrogen of Phe120. <scene name='Sandbox_Reserved_193/His119a_a/1'>His119</scene> then protonates the 5' oxygen on the ribose ring and the transition state falls to form a 2’3’cyclic phosphate intermediate <ref name="Raines" />. | ||
| - | In a secondary and separate reaction, the 2’,3’ cyclic phosphate is hydrolyzed to a mixture of 2'phosphate and 3' hydroxyl. His12 donates a proton to the leaving group of this reaction, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 abstracts the proton from a water molecule, activating it for nucleophilic attack. The activated water molecule attacks the cyclic phosphate causing the cleavage of the 2'3’ cyclic phosphate intermediate. The truncated nucleotide is then released with a 3’ phosphate group <ref name="Raines" />. | + | In a secondary and separate reaction, the 2’,3’ cyclic phosphate is hydrolyzed to a mixture of 2'phosphate and 3' hydroxyl. His12 donates a proton to the leaving group of this reaction, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 abstracts the proton from a water molecule, activating it for nucleophilic attack. The activated water molecule attacks the cyclic phosphate causing the cleavage of the 2'3’ cyclic phosphate intermediate. The truncated nucleotide is then released with a 3’ phosphate group <ref name="Raines" />. |
== '''Ribonuclease A Substrate Binding''' == | == '''Ribonuclease A Substrate Binding''' == | ||
Revision as of 00:05, 9 October 2011
Bovine Pancreatic Ribonuclease A
| |||||||||||
Additional Proteopedia Pages about RNase A
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Raines RT. Ribonuclease A. Chem Rev. 1998 May 7;98(3):1045-1066. PMID:11848924
- ↑ Avey HP, Boles MO, Carlisle CH, Evans SA, Morris SJ, Palmer RA, Woolhouse BA, Shall S. Structure of ribonuclease. Nature. 1967 Feb 11;213(5076):557-62. PMID:6032249
- ↑ Wyckoff HW, Hardman KD, Allewell NM, Inagami T, Johnson LN, Richards FM. The structure of ribonuclease-S at 3.5 A resolution. J Biol Chem. 1967 Sep 10;242(17):3984-8. PMID:6037556
- ↑ Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH Jr, Hardiman O. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat Genet. 2006 Apr;38(4):411-3. Epub 2006 Feb 26. PMID:16501576 doi:10.1038/ng1742
- ↑ 5.0 5.1 5.2 'Lehninger A., Nelson D.N, & Cox M.M. (2008) Lehninger Principles of Biochemistry. W. H. Freeman, fifth edition.'
- ↑ 6.0 6.1 6.2 Wlodawer A, Svensson LA, Sjolin L, Gilliland GL. Structure of phosphate-free ribonuclease A refined at 1.26 A. Biochemistry. 1988 Apr 19;27(8):2705-17. PMID:3401445
- ↑ Birdsall DL, McPherson A. Crystal structure disposition of thymidylic acid tetramer in complex with ribonuclease A. J Biol Chem. 1992 Nov 5;267(31):22230-6. PMID:1429575
- ↑ delCardayre SB, Raines RT. Structural determinants of enzymatic processivity. Biochemistry. 1994 May 24;33(20):6031-7. PMID:8193116
- ↑ Thompson JE, Raines RT. Value of general Acid-base catalysis to ribonuclease a. J Am Chem Soc. 1994 Jun;116(12):5467-8. PMID:21391696 doi:10.1021/ja00091a060
- ↑ Fontecilla-Camps JC, de Llorens R, le Du MH, Cuchillo CM. Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition. J Biol Chem. 1994 Aug 26;269(34):21526-31. PMID:8063789
- ↑ Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995 Mar 9;374(6518):183-6. PMID:7877692 doi:http://dx.doi.org/10.1038/374183a0
- ↑ Turcotte RF, Raines RT. Interaction of onconase with the human ribonuclease inhibitor protein. Biochem Biophys Res Commun. 2008 Dec 12;377(2):512-4. Epub 2008 Oct 16. PMID:18930025 doi:10.1016/j.bbrc.2008.10.032
External Resources
- Full crystal structure of 1RTA
- Full crystal structure of 1RCN
- Further information on 1RTA
- RNase A- Molecule of the Month
- RNase A-Wikipedia
- Ribonuclease Inhibitor-Wikipedia
- Ribonuclease-Wikipedia
- Ruminants-Wikipedia