Retinoblastoma protein
From Proteopedia
| Line 20: | Line 20: | ||
inhibition of E2F and of cyclin CDK complexes. When pRB is phosphorylated, it | inhibition of E2F and of cyclin CDK complexes. When pRB is phosphorylated, it | ||
no longer inhibits cell growth and the cell can continue developing. | no longer inhibits cell growth and the cell can continue developing. | ||
| + | |||
| + | |||
==Specific Binding sites of pRb/E2F complex== | ==Specific Binding sites of pRb/E2F complex== | ||
| - | When pRB and E2F form a complex, pRB is activated, and therefore inhibits cell growth. This complex is held together by many weak interactions, such as hydrophobic and hydrogen bond interactions. The retinoblastoma protein’s active site is composed of two domains. <scene name='Retinoblastoma/E2F_complex/ | + | When pRB and E2F form a complex, pRB is activated, and therefore inhibits cell growth. This complex is held together by many weak interactions, such as hydrophobic and hydrogen bond interactions. The retinoblastoma protein’s active site is composed of two domains. <scene name='Retinoblastoma/E2F_complex/Domains_a_and_b/1'>Domains A and B</scene>, which form the pRB pocket domain that E2F can bind to, the C-terminal residues on pRb are also necessary for E2F to bind with a higher affinity. Specifically, E2F binds to the α4, α5, α6, α8, and α9 helices of the A domain, and only bind to the α11 helix of the B domain (put in figure). This structure was obtained through a diffraction image from a diffractometer (reference). They also found that of the entire E2F complex only the two end regions are needed to bind pRb, the marked box region and the transactivation domain (residues 243-437). |
(highlight the E2F peptide)There are 18 residues of the E2F protein that are involved in the pRb/E2F complex, residues 409-426. Within this E2Fregion, there are 5 essential amino acids that if altered, would cause the complex to fall apart. These amino acids are the following: Tyr(411) binds to pRB via a hydrophobic pocket created by phenolic ring, and a hydrogen bond created by the hydroxyl group on the phenol ring(zoom into that area). Leu(424) and Phe(425) both form many hydrophobic interactions. Glu(419) hydrogen bonds to Thr(645) of pRB, and Aps(423) forms a salt bridge with Arg(467) on pRB (reference). Any point mutations of any of these amino acids will block binding of pRb, leading to deactivation of pRb’s cell inhibiting function, and will allow E2F to promote the cell cycle to move on to the S-phase. | (highlight the E2F peptide)There are 18 residues of the E2F protein that are involved in the pRb/E2F complex, residues 409-426. Within this E2Fregion, there are 5 essential amino acids that if altered, would cause the complex to fall apart. These amino acids are the following: Tyr(411) binds to pRB via a hydrophobic pocket created by phenolic ring, and a hydrogen bond created by the hydroxyl group on the phenol ring(zoom into that area). Leu(424) and Phe(425) both form many hydrophobic interactions. Glu(419) hydrogen bonds to Thr(645) of pRB, and Aps(423) forms a salt bridge with Arg(467) on pRB (reference). Any point mutations of any of these amino acids will block binding of pRb, leading to deactivation of pRb’s cell inhibiting function, and will allow E2F to promote the cell cycle to move on to the S-phase. | ||
Revision as of 17:26, 15 November 2011
| |||||||||
| 1o9k, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Related: | 1pjm, 1n4m, 1h24, 1ad6, 1h25, 1gh6, 1gux, 2aze | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Retinoblastoma Protein (pRB) and E2F
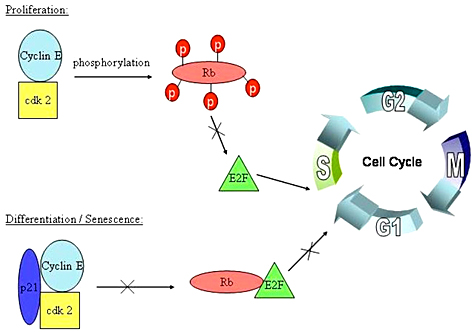
The weighs 110 kDa. It is a tumor suppressor protein that prevents excessive cell growth. If not mutated, it is present in our cells and blocks the growth of dysfunctional cells between the G1 and S phase of the cell cycle before they can undergo DNA replication. pRb also regulates the function of cyclin and cyclin dependent kinase (CDK) complexes which are needed for the cell to proceed with cell growth. E2F is a transcription factor that forms a complex with pRb to inhibit cell growth. When E2F is not bound to pRb it encourages cells to proliferate, it's main function is to express genes required for DNA replication to take place. Retinoblastoma protein is dependent on the transcription factor E2F, which is bound to pRb's active site when it is hypo phosphorylated. In viral cells, and in tumorous cells pRb is dysfunctional or completely lost.If there is a mutation in this gene, it's binding domain is dysfunctional and can lead to excessive cell growth, which may cause tumor or cancer development.
pRb/EF binding in the cell cycle
A cell must go through the cell cycle in order to grow, and replicate. Before the cell can replicate it has to pass through the many phases of the cell cycle. There’s the Go phase, also known as the resting phase in which the cell exits from the cell cycle, and can remain dormant forever. Then comes the G1 phase in which cell growth takes place. Next is the R-phase restriction point which determines if the cell is ready to move on to the S-phase, which is when the cell’s DNA is replicated. It moves on to the G2 phase and finally the M-phase where it divides. The phases where pRB and E2F function are necessary are during the Go phase, and during the restriction point. In these phases pRb will inhibit cell growth, and determine if the cell can transition from the G1 phase to the S phase. If the cell is not growing adequately, for example if its DNA is damaged, then the retinoblastoma protein will hypo-phosphorylate and bind to the E2F transcription factor.This triggers the inhibition of E2F and of cyclin CDK complexes. When pRB is phosphorylated, it no longer inhibits cell growth and the cell can continue developing.
Specific Binding sites of pRb/E2F complex
When pRB and E2F form a complex, pRB is activated, and therefore inhibits cell growth. This complex is held together by many weak interactions, such as hydrophobic and hydrogen bond interactions. The retinoblastoma protein’s active site is composed of two domains. , which form the pRB pocket domain that E2F can bind to, the C-terminal residues on pRb are also necessary for E2F to bind with a higher affinity. Specifically, E2F binds to the α4, α5, α6, α8, and α9 helices of the A domain, and only bind to the α11 helix of the B domain (put in figure). This structure was obtained through a diffraction image from a diffractometer (reference). They also found that of the entire E2F complex only the two end regions are needed to bind pRb, the marked box region and the transactivation domain (residues 243-437).
(highlight the E2F peptide)There are 18 residues of the E2F protein that are involved in the pRb/E2F complex, residues 409-426. Within this E2Fregion, there are 5 essential amino acids that if altered, would cause the complex to fall apart. These amino acids are the following: Tyr(411) binds to pRB via a hydrophobic pocket created by phenolic ring, and a hydrogen bond created by the hydroxyl group on the phenol ring(zoom into that area). Leu(424) and Phe(425) both form many hydrophobic interactions. Glu(419) hydrogen bonds to Thr(645) of pRB, and Aps(423) forms a salt bridge with Arg(467) on pRB (reference). Any point mutations of any of these amino acids will block binding of pRb, leading to deactivation of pRb’s cell inhibiting function, and will allow E2F to promote the cell cycle to move on to the S-phase.
Inhibition/ Activation of pRB
Activation of retinoblastoma protein occurs when it is hypophosphorylated, bound to E2F, and is not mutated. When hypophosphorylated, it inactivates cdk-cyclin complexes and is tightly bound to E2F. A non mutated pRb protein can undergo proper phosphorylation and proper binding to E2F. There are also many ways to inactivate pRb, by phosphorylating it, interaction with viral proteins, mutation of the pRb gene, and via the Caspade-mediated degradation.
Phosphorylation of pRb is caused by binding of CDK-cyclin complexes such as cdk4/6-cyclin D and cdk2-cyclin E (reference black-5). This will prevent binding free pRb to E2F, or can cause the pRb/E2F complex to fall apart. Now that E2F is free, it can activate the progression of the cell though to S-phase. Ser (567) is a specific amino acid that undergoes phosphorylation, and has been observed to disrupt the A and B-domain binding site of pRb (reference Bing Xiao).
When pRb interacts with viral proteins, the viral protein binds to the A and B domains and blocks binding to E2F. Examples of these viral genes are oncoprotein E7 of the Human papillomavirus, the adenovirus E1A which encodes potential oncoproteins that alter cell control, and the SV40 antigen which is a DNA virus that can cause tumors.
Any mutation of the pRb gene will disrupt its proper function. If mutated, pRb can me either present and dysfunctional, or completely absent. Mutated pRb has been found in many human tumors, such as osteosarcoma which is a cancerous bone tumor, cell lung carcinomas and breast carcinomas that arise from cells whose genome has been altered.
The Caspade-Mediated degradation is a type of cleavage that leads to pRb degradation. Lysing occurs between Asp (886) and Gly (887), which then allows a Tumor Nacrosis factor (TNF) to stimulate apoptosis of pRb.
Tumors, cancer and treatments
Disruption of the pRb/E2F complex has been discovered in many human tumors and cancers. Alteration of this complex results in free E2F transcription factor that accumulates in the cell and allows cells to proliferate.
E2F has different roles in various cancers and tumors. Studies done on breast carcinomas, prostate adenocarcinomas, colon adenocarcinomas, and bladder carcinomas, showed that expression of E2F was higher than in regular cells. E2F was found to either allow malignant cells to proliferate or to counterpart with tumor growth.
Studies of the retinoblastoma protein and E2F transcription factor have been done to somehow find a treatment for cancer. A compound, β-Lapachone that comes from trees in South America, is currently used as a drug for cancer treatment. It functions to inhibit proliferation of cells by enhancing the binding of the pRb/E2F complex. It also inhibits the function of cyclin E, and cyclin dependent kinase to block pRb phosphorylation. β-Lapachone also induces cell apoptosis, and reacts with many other transcription factors to stop uncontrollable cell growth The pRb/E2F complex is very important for normal cell growth in our bodies, without proper function of it, humans are prone to the development of tumors, malignant cells, and cancer.
Proteopedia Page Contributors and Editors (what is this?)
Yvonne Alva, Michal Harel, Joel L. Sussman, Alexander Berchansky