A-ATP Synthase
From Proteopedia
| Line 1: | Line 1: | ||
| - | |||
[[Image:F_ATPsynthase.gif | thumb]] | [[Image:F_ATPsynthase.gif | thumb]] | ||
==Structure== | ==Structure== | ||
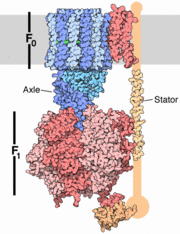
| - | The structure on the right shows the F1 motor and the axle that connects the two. ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor . This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too. | + | The structure on the right shows the F1 motor and the axle that connects the two. ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor . This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too. [http://en.wikipedia.org/wiki/Atp_synthase A-ATP synthase] is very similar to F ATP Synthase and is composed of two parts '''A1''' and '''A0''' which are composed of at least nine subunits '''A3B3C:D:E:F:H2:a:cx''' that function as a pair of rotary motors connected by central and peripheral stalk(s) <ref name= Muller> PMID: 16645313</ref>. The '''A0''' domain is the hydrophobic membrane embedded ion-translocating sector that uses the H+ gradient to power ATP synthase in domain '''A1'''. '''A1''' is catalytic and water soluble containing '''A''' and '''B''' subunits. These subunits are comparable to F-ATP synthase [http://en.wikipedia.org/wiki/ATP_synthase_alpha/beta_subunits ATP synthase alpha/beta subunits]. The '''A''' subunit of '''A1''' is catalytic and the '''B''' subunit is regulatory, with a substrate-binding site on each. [http://www.youtube.com/watch?v=KU-B7G6anqw&feature=related] |
| - | + | ||
| - | that function as a pair of rotary motors connected by central and peripheral stalk(s) <ref name= Muller> PMID: 16645313</ref>. The '''A0''' domain is the hydrophobic membrane embedded ion-translocating sector that uses the H+ gradient to power ATP synthase in domain '''A1'''. '''A1''' is catalytic and water soluble containing '''A''' and '''B''' subunits. These subunits are comparable to F-ATP synthase [http://en.wikipedia.org/wiki/ATP_synthase_alpha/beta_subunits ATP synthase alpha/beta subunits]. The '''A''' subunit of '''A1''' is catalytic and the '''B''' subunit is regulatory, with a substrate-binding site on each. [http://www.youtube.com/watch?v=KU-B7G6anqw&feature=related] | + | |
<StructureSection load=1e79 size='500' side='right' caption='F1-ATP synthase motor domain', ([[1e79]])' scene=''> | <StructureSection load=1e79 size='500' side='right' caption='F1-ATP synthase motor domain', ([[1e79]])' scene=''> | ||
| Line 17: | Line 14: | ||
</StructureSection> | </StructureSection> | ||
| - | |||
| - | |||
| - | |||
<StructureSection load=3p20 size='500' side='right' caption='A-ATP synthase catalytic A subunit', ([[3p20]])' scene=''> | <StructureSection load=3p20 size='500' side='right' caption='A-ATP synthase catalytic A subunit', ([[3p20]])' scene=''> | ||
| Line 48: | Line 42: | ||
==Transient Binding site== | ==Transient Binding site== | ||
| + | |||
Vanadate two The second <scene name='A-ATP_Synthase/1-vandate/1'>vandate</scene> is positioned in a region exactly opposite the nucleotide-binding site, where the ATP molecule transiently associates on its way to the final binding pocket in subunit "'B"'. [L417] Is involved in a bifurcated hydrogen bond with the second vandate. This vanadate is also stabilized by weak non polar interactions with P233 F399 F414 A416 and A419, as well as polar interactions with D418 N431 and T434. Similar binding behavior was observed for "'As"' indicating that the substrate molecule has a similar path of entry to the active site in both the "'A"' and '"B"' subunit of the A-ATP synthase and that they have a transient binding position near the P-Loop. It is proposed that Pi binds first to the catalytic site and sterically hinders ATP binding, thereby selectively allowing binding of ADP. The "'Avi"' structure confirms this, since although both ADP and Vi were present in the crystallized solution, the catalytic A-subunit first permits only the binding of the phosphate analogue Vi. Hence the present "Avi"' structure represents a trapped initial transition state showing for the first time both the entering path and the final Vi-bound state in the catalytic subunit. | Vanadate two The second <scene name='A-ATP_Synthase/1-vandate/1'>vandate</scene> is positioned in a region exactly opposite the nucleotide-binding site, where the ATP molecule transiently associates on its way to the final binding pocket in subunit "'B"'. [L417] Is involved in a bifurcated hydrogen bond with the second vandate. This vanadate is also stabilized by weak non polar interactions with P233 F399 F414 A416 and A419, as well as polar interactions with D418 N431 and T434. Similar binding behavior was observed for "'As"' indicating that the substrate molecule has a similar path of entry to the active site in both the "'A"' and '"B"' subunit of the A-ATP synthase and that they have a transient binding position near the P-Loop. It is proposed that Pi binds first to the catalytic site and sterically hinders ATP binding, thereby selectively allowing binding of ADP. The "'Avi"' structure confirms this, since although both ADP and Vi were present in the crystallized solution, the catalytic A-subunit first permits only the binding of the phosphate analogue Vi. Hence the present "Avi"' structure represents a trapped initial transition state showing for the first time both the entering path and the final Vi-bound state in the catalytic subunit. | ||
| - | |||
| - | |||
==Comparasons to other known structures== | ==Comparasons to other known structures== | ||
| Line 57: | Line 50: | ||
For example, in A-ATP Synthases <scene name='A-ATP_Synthase/P_loop_3p20/12'>F236</scene> is involved in P-Loop stabilization, but its equivalent (alanine) in subunit B of the F-ATP syntheses subunit beta is a key residue in the catalytic process in moving towards the y-phosphate of ATP during catalysis. By comparing the average distances of the alpha carbons of the P-loop residues to the sulfate, vanadate, and PNP molecules, it was found that the PNP molecule is closest, followed by the vanadate then the sulfate. | For example, in A-ATP Synthases <scene name='A-ATP_Synthase/P_loop_3p20/12'>F236</scene> is involved in P-Loop stabilization, but its equivalent (alanine) in subunit B of the F-ATP syntheses subunit beta is a key residue in the catalytic process in moving towards the y-phosphate of ATP during catalysis. By comparing the average distances of the alpha carbons of the P-loop residues to the sulfate, vanadate, and PNP molecules, it was found that the PNP molecule is closest, followed by the vanadate then the sulfate. | ||
| - | |||
In "'F-ATP Synthase"' the homolog to S238 is the non polar A158. Since A158 cannot form hydrogen bonds to interact with the substrate, the P-loop undergoes a conformational change. In A-ATP Synthase the close proximity needed between S238 and the first vandate during transition state is achieved with a hydrogen bond, not a conformational change in the P-loop. | In "'F-ATP Synthase"' the homolog to S238 is the non polar A158. Since A158 cannot form hydrogen bonds to interact with the substrate, the P-loop undergoes a conformational change. In A-ATP Synthase the close proximity needed between S238 and the first vandate during transition state is achieved with a hydrogen bond, not a conformational change in the P-loop. | ||
| Line 64: | Line 56: | ||
</StructureSection> | </StructureSection> | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{STRUCTURE_1e79| PDB=1e79 | SIZE=400| SCENE= |right|CAPTION=Transition state, [[1e79]] }} | {{STRUCTURE_1e79| PDB=1e79 | SIZE=400| SCENE= |right|CAPTION=Transition state, [[1e79]] }} | ||
Revision as of 06:25, 28 November 2011
Structure
The structure on the right shows the F1 motor and the axle that connects the two. ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor . This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too. A-ATP synthase is very similar to F ATP Synthase and is composed of two parts A1 and A0 which are composed of at least nine subunits A3B3C:D:E:F:H2:a:cx that function as a pair of rotary motors connected by central and peripheral stalk(s) [1]. The A0 domain is the hydrophobic membrane embedded ion-translocating sector that uses the H+ gradient to power ATP synthase in domain A1. A1 is catalytic and water soluble containing A and B subunits. These subunits are comparable to F-ATP synthase ATP synthase alpha/beta subunits. The A subunit of A1 is catalytic and the B subunit is regulatory, with a substrate-binding site on each. [1]
| |||||||||||
| |||||||||||
| |||||||
| Transition state, 1e79 | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , , , , | ||||||
| Activity: | H(+)-transporting two-sector ATPase, with EC number 3.6.3.14 | ||||||
| Related: | 1bmf, 1cow, 1e1q, 1e1r, 1efr, 1nbm, 1qo1
| ||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Contents |
References
- ↑ 1.0 1.1 Muller V, Lemker T, Lingl A, Weidner C, Coskun U, Gruber G. Bioenergetics of archaea: ATP synthesis under harsh environmental conditions. J Mol Microbiol Biotechnol. 2005;10(2-4):167-80. PMID:16645313 doi:10.1159/000091563
- ↑ Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000 Nov;7(11):1055-61. PMID:11062563 doi:10.1038/80981
- ↑ Schafer IB, Bailer SM, Duser MG, Borsch M, Bernal RA, Stock D, Gruber G. Crystal structure of the archaeal A1Ao ATP synthase subunit B from Methanosarcina mazei Go1: Implications of nucleotide-binding differences in the major A1Ao subunits A and B. J Mol Biol. 2006 May 5;358(3):725-40. Epub 2006 Mar 10. PMID:16563431 doi:http://dx.doi.org/10.1016/j.jmb.2006.02.057
- ↑ Gonzalez JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998 May;2(2):123-30. PMID:9672687
- ↑ Manimekalai MS, Kumar A, Jeyakanthan J, Gruber G. The Transition-Like State and P(i) Entrance into the Catalytic A Subunit of the Biological Engine A-ATP Synthase. J Mol Biol. 2011 Mar 16. PMID:21396943 doi:10.1016/j.jmb.2011.03.010
- ↑ Priya R, Kumar A, Manimekalai MS, Gruber G. Conserved Glycine Residues in the P-Loop of ATP Synthases Form a Doorframe for Nucleotide Entrance. J Mol Biol. 2011 Sep 8. PMID:21925186 doi:10.1016/j.jmb.2011.08.045
- ↑ Manimekalai MS, Kumar A, Jeyakanthan J, Gruber G. The Transition-Like State and P(i) Entrance into the Catalytic A Subunit of the Biological Engine A-ATP Synthase. J Mol Biol. 2011 Mar 16. PMID:21396943 doi:10.1016/j.jmb.2011.03.010
Proteopedia Page Contributors and Editors (what is this?)
Kaitlin Chase MacCulloch, Michal Harel, Alexander Berchansky