We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Brian Hernandez/DOPA Decarboxylase
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==Structure== | ==Structure== | ||
---- | ---- | ||
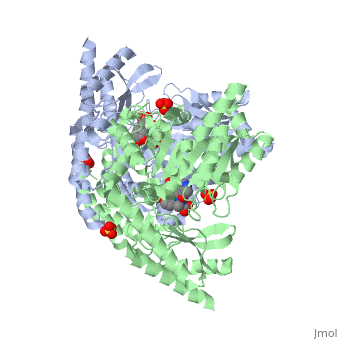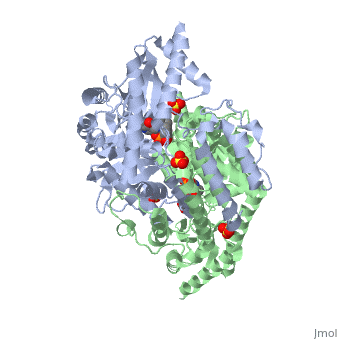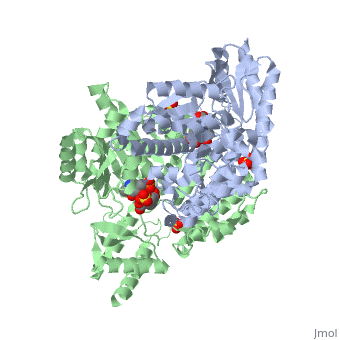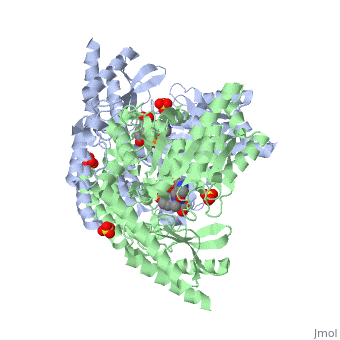
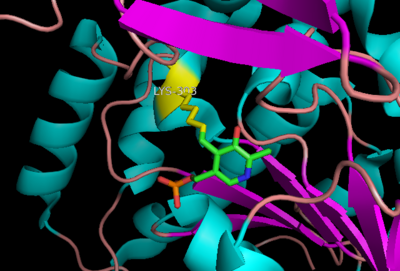
| - | DDC consists of two monomers, each with three distinct domains: the large domain, the C-terminal small domain, and the N-terminal domain <ref name=Burkhard>PMID: 11685243 </ref>. The <scene name='DOPA_decarboxylase/Large_domain/1'>large domain</scene> consists of the cofactor (PLP- pyridoxal phosphate) binding site and is comprised of a central, seven-stranded mixed β-sheet surrounded by eight α-helices in a typical α/β fold. The small <scene name='DOPA_decarboxylase/Small_domain/1'>C-terminal domain</scene> consists of a four-stranded antiparallel β-sheet with three helices packed against the face opposite the large domain. The <scene name='Sandbox/N-terminal_domain/2'>N-terminal domain</scene> (characteristic of all α-family enzymes) is composed of two parallel helices linked by an extended strand. This structure flaps over the top of the second subunit and vice versa, with the first helix of one subunit aligning parallel to the equivalent helix of the other subunit, thereby forming the extended dimer interface. Although it is unlikely that this domain represents an autonomous folding unit, it is most likely stable only in the context of the dimer by extending the interface between the two monomers. | + | DDC consists of two monomers, each with three distinct domains: the large domain, the C-terminal small domain, and the N-terminal domain <ref name=Burkhard>PMID: 11685243 </ref>. The <scene name='DOPA_decarboxylase/Large_domain/1'>large domain</scene> consists of the cofactor (PLP- pyridoxal phosphate) binding site and is comprised of a central, seven-stranded mixed β-sheet surrounded by eight α-helices in a typical α/β fold. The small <scene name='DOPA_decarboxylase/Small_domain/1'>C-terminal domain</scene> consists of a four-stranded antiparallel β-sheet with three helices packed against the face opposite the large domain. The <scene name='Sandbox/N-terminal_domain/2'>N-terminal domain</scene> (characteristic of all α-family enzymes) is composed of two parallel helices linked by an extended strand. This structure flaps over the top of the second subunit and vice versa, with the first helix of one subunit aligning parallel to the equivalent helix of the other subunit, thereby forming the '''extended dimer interface'''. Although it is unlikely that this domain represents an autonomous folding unit, it is most likely stable only in the context of the dimer by extending the interface between the two monomers. |
==Function== | ==Function== | ||
Revision as of 07:01, 29 November 2011
| |||||||||||
3D structures of DOPA decarboxylase
| |||||||||
| 1js3, resolution 2.25Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Activity: | Aromatic-L-amino-acid decarboxylase, with EC number 4.1.1.28 | ||||||||
| Related: | 1js6 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ Christenson JG, Dairman W, Udenfriend S. On the identity of DOPA decarboxylase and 5-hydroxytryptophan decarboxylase (immunological titration-aromatic L-amino acid decarboxylase-serotonin-dopamine-norepinephrine). Proc Natl Acad Sci U S A. 1972 Feb;69(2):343-7. PMID:4536745
- ↑ Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ 3.0 3.1 Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823