This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
ToxT
From Proteopedia
| Line 11: | Line 11: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
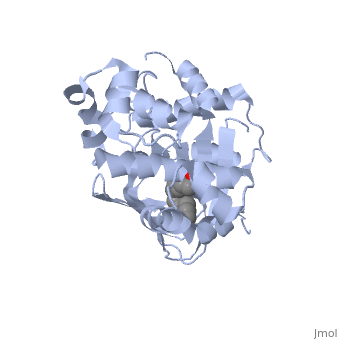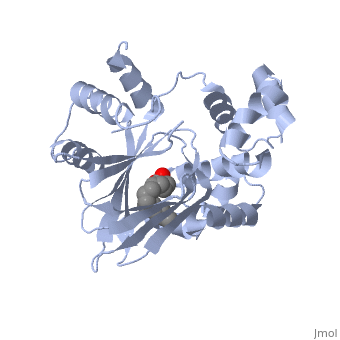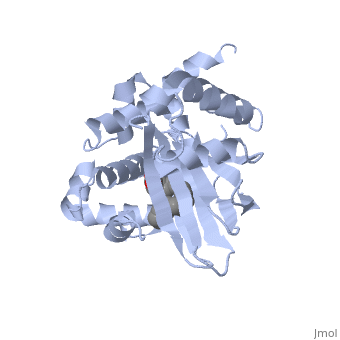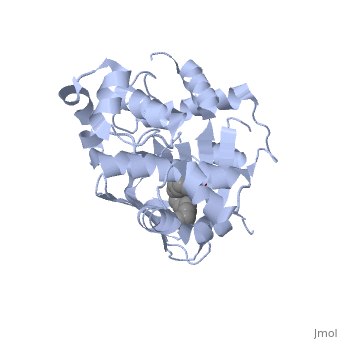
| - | <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Barrel/1'>A nine-stranded beta sheet sandwich</scene> or "jelly-roll" with three other alpha helices (overall making up the <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/N-terminus/1'>N-terminus</scene>) contain a <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/1'>binding pocket</scene>. This is largely made up of residues from the N-terminus (Y12, Y20, F22, L25, I27, K31, F33, L61, F69, L71, V81, and V83), and a few from the C-terminus (I226, K230, M259, V261, Y266, and M269). This pocket contains a ligand: ,<scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/2' | + | <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Barrel/1'>A nine-stranded beta sheet sandwich</scene> or "jelly-roll" with three other alpha helices (overall making up the <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/N-terminus/1'>N-terminus</scene>) contain a <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/1'>binding pocket</scene>. This is largely made up of residues from the N-terminus (Y12, Y20, F22, L25, I27, K31, F33, L61, F69, L71, V81, and V83), and a few from the C-terminus (I226, K230, M259, V261, Y266, and M269). This pocket contains a ligand: ,<scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/2'>cis-palmitoleate</scene> <ref name="structure">PMID: 20133655</ref> which appears to have a negative effect on virulence when present in vitro. This unsaturated fatty acid, like other UFAs,[http://en.wikipedia.org/wiki/Fatty_acid#Unsaturated_fatty_acids] tend to inhibit genes under the control of ToxT. The |
Specifically, the <i>cis</i>-palmitoleate appears to bind directly to ToxT, change its conformation, and thus lower the ability to bind DNA and form dimers.<ref name="structure">PMID: 20133655</ref> | Specifically, the <i>cis</i>-palmitoleate appears to bind directly to ToxT, change its conformation, and thus lower the ability to bind DNA and form dimers.<ref name="structure">PMID: 20133655</ref> | ||
<br/> | <br/> | ||
Revision as of 16:25, 29 November 2011
The crystal structure of ToxT is resolved in monomeric form, after isolation from Vibrio cholerae strain O395.[1]
Contents |
Introduction
ToxT is a molecule at the end of a transcriptional cascade that autoregulates the transcription of the primary virulence factors of Vibrio cholerae[1] and itself. These two factors, cholera toxin (CT)[2] and the toxin co-regulated pilus (TCP), are instrumental in causing the disease cholera[3]. This is an intestinal infection resulting in massive water loss in the affected individual, causing extreme dehydration.[4]
|
Ligand
In this resolved structure, [5] is present and bound in the beta sheet barrel (further discussed below). This unsaturated fatty acid reduces virulence expression in Vibrio cholerae.
Other Structural Features
ToxT belongs to a family of transcriptional regulators headed by and known as AraC.[1] The AraC family is characterized by a 100 amino acid region of sequence similarity that forms a with two helix-turn-helix motifs (helix in blue, turn in teal). [2] The two HTH regions are linked by a very polar alpha helix (shown in black). The overall domain is composed of seven alpha helices, and is located at the C-terminus.[1]
or "jelly-roll" with three other alpha helices (overall making up the ) contain a . This is largely made up of residues from the N-terminus (Y12, Y20, F22, L25, I27, K31, F33, L61, F69, L71, V81, and V83), and a few from the C-terminus (I226, K230, M259, V261, Y266, and M269). This pocket contains a ligand: , [1] which appears to have a negative effect on virulence when present in vitro. This unsaturated fatty acid, like other UFAs,[6] tend to inhibit genes under the control of ToxT. The
Specifically, the cis-palmitoleate appears to bind directly to ToxT, change its conformation, and thus lower the ability to bind DNA and form dimers.[1]
Though the structure shown is a monomer with two overall domains (N-terminal and C-terminal), ToxT tends to form a dimer. The preferred state of ToxT varies between promoters, but binding to the ctx promoter to generate cholera toxin appears to be possible only in the dimer form.[3]ToxT binds to thirteen base pair sequences (can be single, direct, or inverted repeats) called toxboxes in order to activate their respective promoters.[7]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A. 2010 Feb 16;107(7):2860-5. Epub 2010 Feb 1. PMID:20133655
- ↑ Martin RG, Rosner JL. The AraC transcriptional activators. Curr Opin Microbiol. 2001 Apr;4(2):132-7. PMID:11282467
- ↑ Shakhnovich EA, Hung DT, Pierson E, Lee K, Mekalanos JJ. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2372-7. Epub 2007 Feb 5. PMID:17283330 doi:10.1073/pnas.0611643104
Proteopedia Page Contributors and Editors (what is this?)
Ingrid Youngworth, Yang Yang, Michal Harel, Alexander Berchansky, Jaime Prilusky