Forkhead box protein M1
From Proteopedia
m (→See Also) |
|||
| Line 27: | Line 27: | ||
{{Template:ColorKey Bases DNA}} <br\> | {{Template:ColorKey Bases DNA}} <br\> | ||
{{Link Toggle FancyCartoonHighQualityView}} | {{Link Toggle FancyCartoonHighQualityView}} | ||
| + | |||
| + | ==3D structures of forkhead box proteins== | ||
| + | |||
| + | [[Forkhead box protein]] | ||
==Reference== | ==Reference== | ||
| Line 32: | Line 36: | ||
==See Also== | ==See Also== | ||
| - | *[[Forkhead box | + | *[[Forkhead box protein]] |
*[[FOXP2]] | *[[FOXP2]] | ||
*[[Cancer]] | *[[Cancer]] | ||
Revision as of 17:17, 11 January 2012
Learn more about the honor here. |
Contents |
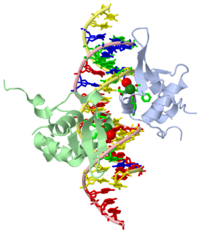
Structure of the FOXM1 DNA binding
FoxM1 is a member of the Forkhead family of transcription factors and is implicated in inducing cell proliferation and some forms of tumorigenesis. It binds promoter regions with a preference for tandem repeats of a consensus 'TAAACA' recognition sequence. The affinity of the isolated FoxM1 DNA-binding domain for this site is in the micromolar range, lower than observed for other Forkhead proteins. To explain these FoxM1 features, we determined the crystal structure of its DNA-binding domain in complex with a tandem recognition sequence. FoxM1 adopts the winged-helix fold, typical of the Forkhead family. Neither 'wing' of the fold however, makes significant contacts with the DNA, while the second, C-terminal, wing adopts an unusual ordered conformation across the back of the molecule. The lack of standard DNA-'wing' interactions may be a reason for FoxM1's relatively low affinity. The role of the 'wings' is possibly undertaken by other FoxM1 regions outside the DBD, that could interact with the target DNA directly or mediate interactions with other binding partners. Finally, we were unable to show a clear preference for tandem consensus site recognition in DNA-binding, transcription activation or bioinformatics analysis; FoxM1's moniker, 'Trident', is not supported by our data.
Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence., Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A, Nucleic Acids Res. 2010 Jul;38(13):4527-38. Epub 2010 Mar 31. PMID:20360045
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
3G73 is a 4 chains structure with sequences from Homo sapiens. Full crystallographic information is available from OCA.
A C G T
3D structures of forkhead box proteins
Reference
- Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010 Jul;38(13):4527-38. Epub 2010 Mar 31. PMID:20360045 doi:10.1093/nar/gkq194
See Also
Original page 3g73 seeded by OCA on Wed Jun 23 08:38:23 2010
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Joel L. Sussman, Michal Harel, Alexander Berchansky
Categories: Homo sapiens | Hibbert, R G. | Littler, D R. | Medema, R H. | Perrakis, A. | Activator | Alternative splicing | Dna-binding | Dna-binding domain | Forkhead | Forkhead transcription factor | Foxm1 | Nucleus | Phosphoprotein | Polymorphism | Transcription | Transcription regulation | Transcription-dna complex | Winged helix | Topic Page