Sandbox 39
From Proteopedia
| Line 37: | Line 37: | ||
== Primary, Secondary, and Tertiary Structure == | == Primary, Secondary, and Tertiary Structure == | ||
| - | Papain is composed of 212 amino acid residues that make up its primary structure. | + | Papain is composed of 212 amino acid residues that make up its primary structure. |
| - | + | ||
| - | + | ||
Papain's primary structure causes it to fold into different motifs that make up its secondary structure. These motifs include <scene name='Sandbox_39/Alpha_helices/2'>alpha helices</scene>, shown in green, and <scene name='Sandbox_39/Beta_pleated_sheets/2'>beta pleated sheets</scene>, shown in orange. As shown to the left, papain has 7 alpha helices and 8 beta pleated sheets. All other motifs are nonrandom, structural units, mostly simply turns. | Papain's primary structure causes it to fold into different motifs that make up its secondary structure. These motifs include <scene name='Sandbox_39/Alpha_helices/2'>alpha helices</scene>, shown in green, and <scene name='Sandbox_39/Beta_pleated_sheets/2'>beta pleated sheets</scene>, shown in orange. As shown to the left, papain has 7 alpha helices and 8 beta pleated sheets. All other motifs are nonrandom, structural units, mostly simply turns. | ||
Papain's seconary structures then fold even further to for its three-dimensional structure, which consists of two distinct structural domains with a cleft between them. This cleft contains the catalytic diad discussed above. This tertiary structure is pictured to the right. | Papain's seconary structures then fold even further to for its three-dimensional structure, which consists of two distinct structural domains with a cleft between them. This cleft contains the catalytic diad discussed above. This tertiary structure is pictured to the right. | ||
| - | |||
[[Image:9pap moreau 1.jpeg|right]] | [[Image:9pap moreau 1.jpeg|right]] | ||
<ref>Image from: | <ref>Image from: | ||
| Line 95: | Line 92: | ||
3. Hans-Hartwig Otto, and Tanja Schirmeister (1997) Cysteine Proteases and Their Inhibitors. Chemical Reviews. No. 97, 133-171. | 3. Hans-Hartwig Otto, and Tanja Schirmeister (1997) Cysteine Proteases and Their Inhibitors. Chemical Reviews. No. 97, 133-171. | ||
| - | 4 | + | 4. Image from: http://linus.chem.ku.edu/hewlett/Chem188/Enzyme/enzyme_background.htm |
| - | + | ||
| - | + | ||
| - | + | 5. Image from: http://www.rcsb.org/pdb/explore/jmol.do?structureId=9PAP&bionumber=1 | |
| - | + | 6. Image from: http://www.rcsb.org/pdb/explore/explore.do?structureId=1POP | |
Revision as of 19:46, 8 March 2012
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
Papain
|
Introduction
Papain (9PAP), also known as papaya proteinase I, is an enzyme found in unripe papaya fruit. A cysteine protease, it has been used to break down tough muscle fibers, and hence is often found in powdered meat tenderizers. It has also been used in cell isolation procedures because it is very efficient and not very destructive. It is collected from the fruit by scoring its skin and allowing the "sap" to seem out. The sap is then dried and purified.
The 3D model of papain shown to the right is "decorated" with solvent methanol molecules. The following Jmol representations of papain will not show these solvent molecules.
Function
As a cysteine protease, Papain utilizes a nucleophilic cysteine thiol as part of its catalytic triad. Papain's Cys-25 is deprotonated by its His-159. The now nucleophilic Cys-25 attacks the carbonyl carbon of the peptide backbone, forming an acyl enzyme intermediate in which the peptide's amino terminal is free. Also in this step, His-159 is returned to its deprotonated form. The intermediate is then deacylated by a water molecule, and it releases the carboxyl terminal of the peptide to produce the product and regenerate the active enzyme. This entire mechanism is shown below:
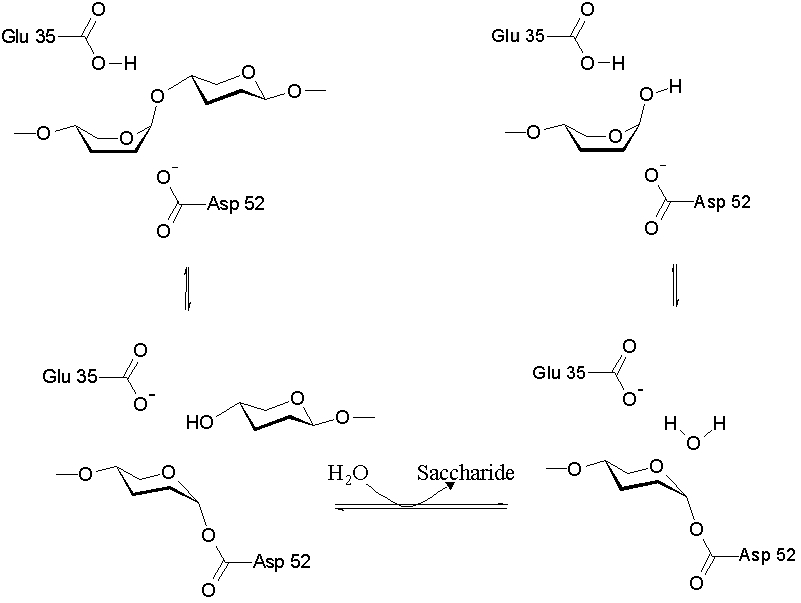 [1]]]
[1]]]
Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains.
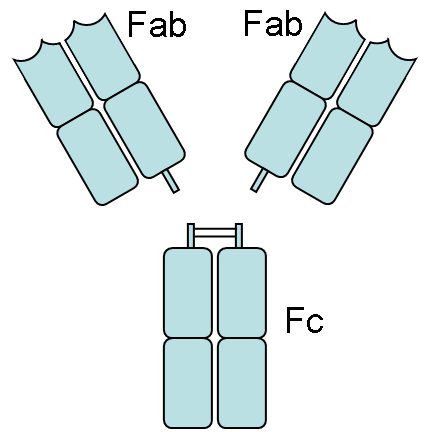
Papain is also known to cleave antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment, as shown below:
[2]]]
|
Composition of Papain
Proteins only consist of certain elements: carbon, hydrogen, nitrogen, oxygen, and sulfur. Enzymes' primary structures allow them to fold optimally and interact with their substrates maximally in order to efficiently catalyze biological reactions. The of papain, seen to the right as a space-fill model, shows carbon atoms outlined in grey, oxygen atoms in red, nitrogen atoms in blue, and sulfur atoms in yellow. In addition, the entirety of the secondary structure of papain can be traced from the As shown to the left, the red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the blue end at the C-terminus.
Primary, Secondary, and Tertiary Structure
Papain is composed of 212 amino acid residues that make up its primary structure. Papain's primary structure causes it to fold into different motifs that make up its secondary structure. These motifs include , shown in green, and , shown in orange. As shown to the left, papain has 7 alpha helices and 8 beta pleated sheets. All other motifs are nonrandom, structural units, mostly simply turns.
Papain's seconary structures then fold even further to for its three-dimensional structure, which consists of two distinct structural domains with a cleft between them. This cleft contains the catalytic diad discussed above. This tertiary structure is pictured to the right.
[3]]]
Active Site
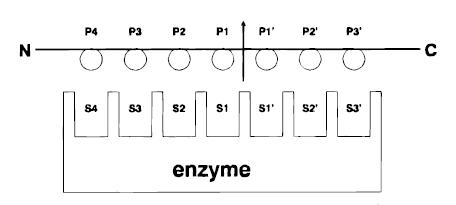
Papain has a broad specificity for protein substrates. The active site consists of seven subsites (S1-S4 and S1’-S3’) that can each accommodate one amino acid residue of a protein substrate (P1-P4 and P1’-P3’).
[4]]] Specificity is controlled, however, by the , a hydrophobic pocket that accommodates the side chains of the protein substrate. This diad, shown above in red and clearly visible in the cleft of the enzyme, consists of cysteine25 (after which the protein is categorized as a cysteine protease)and histidine159. Papain exhibits specific substrate preferences for hydrophobic or aromatic residues.
Polarity and Hydrophobicity
Papain contains many hydrophobic, or "water-hating" regions, and hydrophillic, or "water-loving" regions. The hydrophobic effect, or the tendency of nonpolar substances to aggregate in aqueous solution and exclude water molecules, allows proteins to fold the way they do, exposing hydrophilic residues on their outer surface while sequestering hydrophobic residues in their center. As shown above, all of the are found closer to the center of the folded protein, shown at the left in pink, while the are found around the outside of the protein, shown in blue. These spacefill models give a better idea of the surface area of the protein that contacts the aqueous solution.
Disulfide Bonds
Papain contains three . These bonds are found between Cys-22 and Cys 63, between Cys-56 and Cys-95, and between Cys-153 and Cys-200. These bonds can be seen above as yellow rods, connecting Cysteine residues, also shown in yellow.
Hydrogen Bonding
Hydrogen bonds are essential for the stability of a protein. In the image shown below, the hydrogen bonds can be seen between the secondary structures of the protein as white, dashed lines. Papain contains very specific hydrogen bonding between the amino acid residues. This representation clearly shows how crucial hydrogen bonding is to help maintain the stability of the protein.
Papain Inhibition
|
Substances that inhibit enzymes have sequences that resemble the normal substrate of that enzyme. Some substances that act to inhibit the enzymatic activity of papain are able to do so because of their structural and chemical similarity to polypeptides normally degraded by papain. One example of a papain inhibitor is cystatin. According to this model, the N terminal of the cystatin interacts with the active site and the S1-S3 sites of papain. At the same time, two hairpin loops bind to the S1’-S2’ sites. The interaction between systatin and papain can be seen below. The inactivation of the cysteine proteases, including papain, occurs by competitive, noncovalent, reversible inhibition.
Another example of a papain inhibitor is , a diazomethylketone inhibitor. As shown in the Jmol to the right, the methylene carbon atom of the inhibitor (shown as a grey sphere), is covalently bound to the Cys-25 of papain. The hydrophobic S2 pocket is occupied by the inhibitor's P2 side chain, shown as a pink chain. Extensive hydrogen bonding and hydrophobic interactions are responsible for the interaction of the inhibitor with the enzyme.
Several other molecules have been shown to have protease inhibitory actions. Leupeptin is a naturally-occuring, microbial protease inhibitor. Shown below, sitting in the active site of papain, this molecule contains an arginine residue at its C-terminus that is essential for its inhibitory action. Antipain, a protease specific to papain and trypsin, is a microbial product isolated from actinomycetes. This inhibitor contains a catalytic aldehyde and acts like leupeptin. Antipain is often used in typical protease inhibitor cocktails. Image:Leupeptin.jpg [6]]]
References
1. Image from: http://upload.wikimedia.org/wikipedia/commons/5/5c/Cysteinprotease_Reaktionsmechanismus.svg
2. Image from: http://en.wikipedia.org/wiki/Papain
3. Hans-Hartwig Otto, and Tanja Schirmeister (1997) Cysteine Proteases and Their Inhibitors. Chemical Reviews. No. 97, 133-171.
4. Image from: http://linus.chem.ku.edu/hewlett/Chem188/Enzyme/enzyme_background.htm
5. Image from: http://www.rcsb.org/pdb/explore/jmol.do?structureId=9PAP&bionumber=1
6. Image from: http://www.rcsb.org/pdb/explore/explore.do?structureId=1POP