Sandbox 34
From Proteopedia
| Line 20: | Line 20: | ||
=== Solvent Interactions === | === Solvent Interactions === | ||
Papain binds both <scene name='Sandbox_34/Papainwithwaterandmtoh/1'>methanol and water molecules</scene> via hydrogen bonding that give stability to the papain crystalline structure. This solvent mixture of 62%, (w/w) methanol to water was used in order to obtain a C-type crystal. Papain has an interesting method of utilizing hydrogen bonding with <scene name='Sandbox_34/Papainwithwateronly/1'>water molecules</scene>, shown in cyan. A refined crystal structure of papain revealed that water forms something similar to a hydration shell around individual molecules of papain. The interaction of papain with these water molecules leads to less interaction between papain molecules. The water molecules also form hydrogen bonds within the <scene name='Sandbox_34/Papainwithwaterandmtoh/2'>active site</scene>, providing even more stability for the structure.<ref name="Structure" /> | Papain binds both <scene name='Sandbox_34/Papainwithwaterandmtoh/1'>methanol and water molecules</scene> via hydrogen bonding that give stability to the papain crystalline structure. This solvent mixture of 62%, (w/w) methanol to water was used in order to obtain a C-type crystal. Papain has an interesting method of utilizing hydrogen bonding with <scene name='Sandbox_34/Papainwithwateronly/1'>water molecules</scene>, shown in cyan. A refined crystal structure of papain revealed that water forms something similar to a hydration shell around individual molecules of papain. The interaction of papain with these water molecules leads to less interaction between papain molecules. The water molecules also form hydrogen bonds within the <scene name='Sandbox_34/Papainwithwaterandmtoh/2'>active site</scene>, providing even more stability for the structure.<ref name="Structure" /> | ||
| - | |||
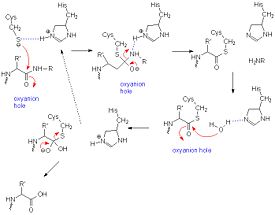
[[Image:Papainmech6.jpg|275px|right|thumb| A general mechanism of papain catalysis<ref>[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref>.]] | [[Image:Papainmech6.jpg|275px|right|thumb| A general mechanism of papain catalysis<ref>[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref>.]] | ||
Revision as of 22:09, 10 March 2012
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
Papain
|
Introduction
Papain is a cysteine protease, also known as papaya proteinase I, from the peptidase C1 family (E.C. 3.4.22.2).[1] It functions as an endopeptidase, amidase, and esterase,[2] with its optimal activity values for pH lying between 6.0 and 7.0, and its optimal temperature for activity is 65 °C. Its pI values are 8.75 and 9.55, and it is best visualized at a wavelength of 278 nm.[3] While only consisting of a single peptide chain, papain has that form a cleft in which the lies.[4] Naturally found in the latex of the papaya fruit, one of the most common uses of papain is as a meat tenderizer because of its ability to hydrolyze esters and amides.[5] Another common use is as a digestive aid. Papaya is commonly referenced as a preferred fruit for those suffering from gastroesophageal reflux disease due to its ability to help the the stomach with digestion of complex proteins.
History
Papain's enzymatic use was first discovered in 1873 by G.C. Roy who published his results in the Calcutta Medical Journal in the article, "The Solvent Action of Papaya Juice on Nitrogenous Articles of Food." In 1879, papain was named officially by Wurtz and Bouchut, who managed to partially purify the product from the sap of papaya. It wasn't until the mid-twentieth century that the complete purification and isolation of papain was achieved. In 1968, Drenth et al. determined the structure of papain by x-ray crystallography, making it the second enzyme whose structure was successfully determined by x-ray crystallography. Additionally, papain was the first cysteine protease to have its structure identified.[2] In 1984, Kamphuis et al. determined the geometry of the active site, and the three-dimensional structure was visualized to a 1.65 Angstrom solution.[6] Today, studies continue on the stability of papain, involving changes in environmental conditions as well as testing of inhibitors such as phenylmethanesulfonylfluoride (PMSF), TLCK, TPCK, aplh2-macroglobulin, heavy metals, AEBSF, antipain, cystatin, E-64, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents.[2]
Structure
|
Papain is a relatively simple enzyme. It consists of only one chain of 212 residues, with a secondary structure composed of 21% and 25% .[7] Its tertiary structure has three disulfide bonds, illustrated in yellow, between Cys22 and Cys63, Cys56, and Cys 95,and Cys153 and Cys200. The single chain is separated into : R in purple, and L in gray. A cleft is formed in which the , consisting of Cys25, His159, and Asp175, resides.[4] A illustration of this shows, from the N-terminus in blue to the C-terminus in red, an easy means by which to track the residues through the molecule. Many hydrogen bonds, illustrated in white, between both the and the help coordinate papain's 3D conformation. also strongly contribute to the stability of the protein structure. In this particular image, clarification of residue coordination is demonstrated by color: paired residues are shown in the same color, oxygen is shown in red, and nitrogen is shown in blue A modified cysteine residue with a sulfhydryl group, , is necessary for the activity of the enzyme[8] In 9PAP, the primary representation of papain used in this article, the sulfhydryl group has been oxidized. Papain contains many . The are illustrated in gray, and the are illustrated in magenta. In a paper entitled, The Structure of Papain Refined at 1.65 A Resoltion, Kamphuis et al. discovered interesting information on between molecules of papain in solution. These contacts, communicated in Table 7 of their paper, consist of nine hydrogen-bond connections and three ionic interactions. The strongest salt bridge exists between .[6]
Solvent Interactions
Papain binds both via hydrogen bonding that give stability to the papain crystalline structure. This solvent mixture of 62%, (w/w) methanol to water was used in order to obtain a C-type crystal. Papain has an interesting method of utilizing hydrogen bonding with , shown in cyan. A refined crystal structure of papain revealed that water forms something similar to a hydration shell around individual molecules of papain. The interaction of papain with these water molecules leads to less interaction between papain molecules. The water molecules also form hydrogen bonds within the , providing even more stability for the structure.[6]
Specificity
Functioning as either an endopeptidase, an amidase, or an esterase, papain functions with a very broad specificity.[10]. It prefers amino acids that bear large hydrophobic side chains at the P2 position, and will not accept valine at the P1' position. [1] Given its broad specificity, papain serves as a cheap and available cysteine protease which can be readily utilized by researchers as a prime example of the mechanisms of inhibition of other enzymes within the cysteine protease super-family.
Catalytic Mechanism
The mechanism of cysteine proteases is very similar to that of serine proteases. However, instead of requiring a triad, papain only requires a diad. The sulfhydryl group on cysteine executes a nucleophilic attack on the peptide bond of the protein it wishes to cleave. Asparagine-175 keeps histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual mechanism. Opening up the carbonyl, the sulfhydryl group of CYS-25 is stabilized by HIS-159. As the carbonyl reforms, the peptide bond is broken, leaving the amide group to fend for itself.[11]
Inhibitors
There are many inhibitors for papain because of its broad specificity. It is often used as a model enzyme for those in the papain super-family, such as cathepsin L and cathepsin K. The inhibition of papain is usually due to active site restriction of cysteine-25 and histidine-159. The interest in developing inhibitors for the papain super-family lies with the desire to effectively inhibit other cysteine proteases that incur unfavorable effects within the body.
|
Cathepsin L
is an endosomal cysteine protease that is believed to have both physiological and pathophysiological effects on the human body. It has been indicated not only in cancer, rhematoid arthritis, and osteo-arthritis, but its mechanism also appears similar to that of Ebola, SARS, and Leishmania. Understanding the mechanism of inhibition through the use of papain is therefore crucial to developing treatments for such diseases.[12] An interesting inhibitor for cathepsin L developed using papain as the model protease is that of .[13] It forms a with Cys25. Five other residues are also involved in the bonding of Clik-148 to papain: Gln19, Gly66, Asp158, Trp177, and Ser205. These participate in hydrophobic, , and hydrogen bonding that effectively fill up the cleft between the two domains of papain.[14]
Cathepsin K
The goal of research for the development of an inhibitor for is the hope to develop a treatment for osteoporosis. In two different cathepsin K inhibitors, referenced PDB codes and , it is evident that the inhibitor binds with much closer proximity than that of Clik148. 1BP4 is a cathepsin K inhibitor, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, that inhibits by interacting with 11 different residues on papain: Gln19, Gly20, Ser21, Gly23, Asn64, Gly65, Gln142, Asp158, His159, Trp177, and Trp181. These interactions range from hydrophobic, electrostatic, and hydrogen bonding, to between the aromatic ring of the carbobenzyl group on 1BP4, and TRP177 of papain. The inhibition of papain by IBQI, carbobenzyloxy-(L)-leucinyl-(L)leucinyl methoxymethylketone, is quite similar to that of IBP4, although it does not bind quite as tightly. It binds to seven residues of papain: Gln19, Gly23, Gly65, Gln142, His159, Trp177, Trp181. Additionally, it has similar between the Cbz ring on the inhibitor and Trp 177, though it is more difficult to visualize with the given PDB file.[15]
Human Stefin B
Other inhibitors, such as , illustrated in magenta, are much more complex in their . The human stefin B molecule has a five stranded beta-sheet that wraps around a five turn alpha-helix. The interface between human stefin B and papain is very tightly packed with 16% of stefin B becoming embedded within papain. A total of 128 <4 A occur within the cleft in papain, although only CYS25 interacts with the inhibitor. In this figure, the residues of interaction for stefin B are shown in red, the residues of interaction for papain are shown in blue, and the residues of papain's active site are shown in green. Through this study, Stubbs et al. were able to conclude that cysteine proteinase inhibitors are "fundamentally different to [those] observed for serine proteinase inhibitors."[16]
References
- ↑ 1.0 1.1 http://www.uniprot.org/uniprot/P00784
- ↑ 2.0 2.1 2.2 http://www.worthington-biochem.com/pap/default.html
- ↑ http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzymes/papain.html
- ↑ 4.0 4.1 http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=9pap&template=clefts.html&r=speedfill
- ↑ IUBMB Enzyme Nomenclature: www.chem.qmul.ac.uk/iubmb/enzyme/EC3/4/22/2.html
- ↑ 6.0 6.1 6.2 Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233-56. PMID:6502713
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=9PAP
- ↑ http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzymes/papain.html
- ↑ [1] University of Maine
- ↑ http://www.ebi.ac.uk/QuickGO/GTerm?id=GO:0004197
- ↑ [2] University of Maine
- ↑ Myers MC, Shah PP, Beavers MP, Napper AD, Diamond SL, Smith AB 3rd, Huryn DM. Design, synthesis, and evaluation of inhibitors of cathepsin L: Exploiting a unique thiocarbazate chemotype. Bioorg Med Chem Lett. 2008 Jun 15;18(12):3646-51. Epub 2008 May 1. PMID:18499453 doi:10.1016/j.bmcl.2008.04.065
- ↑ Tsuge H, Nishimura T, Tada Y, Asao T, Turk D, Turk V, Katunuma N. Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain-CLIK148 complex. Biochem Biophys Res Commun. 1999 Dec 20;266(2):411-6. PMID:10600517 doi:10.1006/bbrc.1999.1830
- ↑ Beavers MP, Myers MC, Shah PP, Purvis JE, Diamond SL, Cooperman BS, Huryn DM, Smith AB 3rd. Molecular docking of cathepsin L inhibitors in the binding site of papain. J Chem Inf Model. 2008 Jul;48(7):1464-72. Epub 2008 Jul 4. PMID:18598021 doi:10.1021/ci800085c
- ↑ LaLonde JM, Zhao B, Smith WW, Janson CA, DesJarlais RL, Tomaszek TA, Carr TJ, Thompson SK, Oh HJ, Yamashita DS, Veber DF, Abdel-Meguid SS. Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites. J Med Chem. 1998 Nov 5;41(23):4567-76. PMID:9804696 doi:10.1021/jm980249f
- ↑ Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990 Jun;9(6):1939-47. PMID:2347312
