Sandbox 22
From Proteopedia
| Line 20: | Line 20: | ||
[[Image:Domains of Papain.png|200px|left|thumb|Contacts between Papain Subunits.<ref name="Richardson">[http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2i.html] Jane S. Richardson</ref>.]] | [[Image:Domains of Papain.png|200px|left|thumb|Contacts between Papain Subunits.<ref name="Richardson">[http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2i.html] Jane S. Richardson</ref>.]] | ||
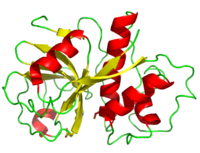
Papain is a relatively simple enzyme, consisting of a single 212 residue chain. A majority of papain's residues, shown in purple in the link, are <scene name='Sandbox_35/Hydrophobicity_papain/3'>hydrophobic</scene>. As with all proteins, it is primarily the exclusion of these residues by water that leads to papain's assumption of a globular form. Despite its apparent simplicity and small size, papain folds into two distinct, evenly sized <scene name='Sandbox_36/Papain_domains/2'>domains</scene>, each with its own <scene name='Sandbox_35/Nonpolar_papain/2'>hydrophobic core</scene>. | Papain is a relatively simple enzyme, consisting of a single 212 residue chain. A majority of papain's residues, shown in purple in the link, are <scene name='Sandbox_35/Hydrophobicity_papain/3'>hydrophobic</scene>. As with all proteins, it is primarily the exclusion of these residues by water that leads to papain's assumption of a globular form. Despite its apparent simplicity and small size, papain folds into two distinct, evenly sized <scene name='Sandbox_36/Papain_domains/2'>domains</scene>, each with its own <scene name='Sandbox_35/Nonpolar_papain/2'>hydrophobic core</scene>. | ||
| - | These two subunits are held together with "arm" linkage where one end of the protein chain holds the opposite domain. In papain's case the "arm crossing" primarily occurs on or near the surface.<ref name="Richardson" /> It is between these two domains that the | + | These two subunits are held together with "arm" linkage, pictured to the left, where one end of the protein chain holds the opposite domain. In papain's case the "arm crossing" primarily occurs on or near the surface.<ref name="Richardson" /> It is between these two domains that the <scene name='Sandbox_22/9pap_bindingpocket_wrtdomains/1'>substrate binding pocket</scene> is situated.<ref>http://books.google.com/books?hl=en&lr=&id=fk1hbZdPTEgC&oi=fnd&pg=PA79&dq=aromatic+residues+in+papain&ots=L8SvlkQaZU&sig=xZ2l8kj52PD7DzuiAQ1zah0CU2M#v=onepage&q=aromatic%20residues%20in%20papain&f=false</ref> The remaining residues are <scene name='Sandbox_36/Papain_polar_residues/1'>polar</scene>, some carrying a <scene name='Sandbox_36/Papain_polar_residues_acidic/1'>negative charge</scene> (acidic) at physiological pH, others a <scene name='Sandbox_36/Papain_polar_residues_basic/1'>positive charge</scene> (basic), the rest of the polar residues are neutral. As expected, the charged <scene name='Sandbox_31/Termini/4'>Termini</scene> face outward due to their hydrophilic nature. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Papain's secondary structure is composed of 21% <scene name='Sandbox_34/Betasheets/1'>beta sheets</scene> (45 residues comprising 7 sheets) and 25% <scene name='Sandbox_34/Alphahelices/1'>alpha helices</scene> (51 residues comprising 7 helices). The rest of the residues, accounting for over 50% of the enzymes structure, make up ordered non-repetative sequences.<ref name="RSCB PDB">http://www.rcsb.org/pdb/explore/explore.do?structureId=9PAP</ref> These secondary structures may be traced from the N- to C-terminus by means of <scene name='Sandbox_39/Elemental/3'>differential coloration</scene>. As shown in this scene, the red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the blue end at the C-terminus. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the <scene name='9pap/Papain_sam_centralhelix/1'>first helix</scene> (residues 25-42) is maintained as a result of <scene name='9pap/Papain_sb_helix1_hbonds/1'>hydrogen bonds</scene> between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, <scene name='9pap/Papain_sb_helix1_hydrophobic/1'>no hydrogen bonds</scene> are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many <scene name='Sandbox_30/Papain_secondary_helix1_phobic/1'>hydrophobic residues</scene> (red residues are hydrophilic). | Papain's secondary structure is composed of 21% <scene name='Sandbox_34/Betasheets/1'>beta sheets</scene> (45 residues comprising 7 sheets) and 25% <scene name='Sandbox_34/Alphahelices/1'>alpha helices</scene> (51 residues comprising 7 helices). The rest of the residues, accounting for over 50% of the enzymes structure, make up ordered non-repetative sequences.<ref name="RSCB PDB">http://www.rcsb.org/pdb/explore/explore.do?structureId=9PAP</ref> These secondary structures may be traced from the N- to C-terminus by means of <scene name='Sandbox_39/Elemental/3'>differential coloration</scene>. As shown in this scene, the red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the blue end at the C-terminus. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the <scene name='9pap/Papain_sam_centralhelix/1'>first helix</scene> (residues 25-42) is maintained as a result of <scene name='9pap/Papain_sb_helix1_hbonds/1'>hydrogen bonds</scene> between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, <scene name='9pap/Papain_sb_helix1_hydrophobic/1'>no hydrogen bonds</scene> are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many <scene name='Sandbox_30/Papain_secondary_helix1_phobic/1'>hydrophobic residues</scene> (red residues are hydrophilic). | ||
<scene name='Sandbox_34/Salt_bridges/5'>Salt bridges</scene> strongly contribute to the tertiary structure of papain. In this particular image, clarification of residue coordination is demonstrated by color: paired residues are shown in the same color, oxygen is shown in red, and nitrogen is shown in blue. The tertiary structure of papain is also maintained by three <scene name='9pap/9pap_sam_disulfides/1'>disulfide bonds</scene>, which connect <scene name='9pap/9pap_sam_disulfides_22-63/1'>Cys-22 to Cys63</scene>, <scene name='9pap/9pap_sam_disulfides_56-95/1'>Cys-56 to Cys-95</scene>, and <scene name='9pap/9pap_sam_disulfides_153-200/1'>Cys-153 to Cys-200</scene><ref name="9PAP PDB" />. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures. | <scene name='Sandbox_34/Salt_bridges/5'>Salt bridges</scene> strongly contribute to the tertiary structure of papain. In this particular image, clarification of residue coordination is demonstrated by color: paired residues are shown in the same color, oxygen is shown in red, and nitrogen is shown in blue. The tertiary structure of papain is also maintained by three <scene name='9pap/9pap_sam_disulfides/1'>disulfide bonds</scene>, which connect <scene name='9pap/9pap_sam_disulfides_22-63/1'>Cys-22 to Cys63</scene>, <scene name='9pap/9pap_sam_disulfides_56-95/1'>Cys-56 to Cys-95</scene>, and <scene name='9pap/9pap_sam_disulfides_153-200/1'>Cys-153 to Cys-200</scene><ref name="9PAP PDB" />. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures. | ||
Revision as of 23:00, 27 March 2012
This sandbox is currently being used to develop a page for papain. Please do not edit it. Thanks, Samuel Bray
Contents |
Papain
Exciting Introduction!
Papain. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings[1]. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, Carica papaya and Vasconcellea cundinamarcensis would have such a practical application beyond proteopedia?
Introduction
General Characteristics
Papain is a 23.4 kDa, 212 residue cysteine protease, also known as papaya proteinase I, from the peptidase C1 family (E.C. 3.4.22.2).[2][3] It is the natural product of the Papaya(Carica papaya)[4], and may be extracted from the plant's latex, leaves and roots.[5] Papain displays a broad range of functions, acting as an endopeptidase, amidase, and esterase,[6] with its optimal activity values for pH lying between 6.0 and 7.0, and its optimal temperature for activity is 65 °C. Its pI values are 8.75 and 9.55, and it is best visualized at a wavelength of 278 nm.[7]
Common Uses
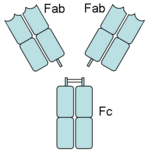
Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains. This non-specific nature of papain's hydrolase activity has led to its use in many and varied commercial products. It is often used as a meat tenderizer because it can hydrolyze the peptide bonds of collagen, elastin, and actomyosin. It is also used in contact lens solution to remove protein deposits on the lenses and marketed as a digestive supplement. Another place papain has found use is clinical medicine, where it is used to treat pain, swelling, and fluid retention following trauma and surgery. [8] Finally, papain has several notable uses in general biomedical research, including a gentle cell isolation agent, production of glycopeptides from purified proteoglycans, and solubilization of integral membrane proteins. It is also notable for its ability to specifically cleave IgG and IgM antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment, as shown in the image to the right:[6]
History
Papain's enzymatic use was first discovered in 1873 by G.C. Roy who published his results in the Calcutta Medical Journal in the article, "The Solvent Action of Papaya Juice on Nitrogenous Articles of Food." In 1879, papain was named officially by Wurtz and Bouchut, who managed to partially purify the product from the sap of papaya. It wasn't until the mid-twentieth century that the complete purification and isolation of papain was achieved. In 1968, Drenth et al. determined the structure of papain by x-ray crystallography, making it the second enzyme whose structure was successfully determined by x-ray crystallography. Additionally, papain was the first cysteine protease to have its structure identified.[6] In 1984, Kamphuis et al. determined the geometry of the active site, and the three-dimensional structure was visualized to a 1.65 Angstrom solution.[9] Today, studies continue on the stability of papain, involving changes in environmental conditions as well as testing of inhibitors such as phenylmethanesulfonylfluoride (PMSF), TLCK, TPCK, aplh2-macroglobulin, heavy metals, AEBSF, antipain, cystatin, E-64, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents.[6]
Structure
|
General Structural Features
Papain is a relatively simple enzyme, consisting of a single 212 residue chain. A majority of papain's residues, shown in purple in the link, are . As with all proteins, it is primarily the exclusion of these residues by water that leads to papain's assumption of a globular form. Despite its apparent simplicity and small size, papain folds into two distinct, evenly sized , each with its own . These two subunits are held together with "arm" linkage, pictured to the left, where one end of the protein chain holds the opposite domain. In papain's case the "arm crossing" primarily occurs on or near the surface.[10] It is between these two domains that the is situated.[11] The remaining residues are , some carrying a (acidic) at physiological pH, others a (basic), the rest of the polar residues are neutral. As expected, the charged face outward due to their hydrophilic nature.
Papain's secondary structure is composed of 21% (45 residues comprising 7 sheets) and 25% (51 residues comprising 7 helices). The rest of the residues, accounting for over 50% of the enzymes structure, make up ordered non-repetative sequences.[12] These secondary structures may be traced from the N- to C-terminus by means of . As shown in this scene, the red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the blue end at the C-terminus. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the (residues 25-42) is maintained as a result of between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many (red residues are hydrophilic).
strongly contribute to the tertiary structure of papain. In this particular image, clarification of residue coordination is demonstrated by color: paired residues are shown in the same color, oxygen is shown in red, and nitrogen is shown in blue. The tertiary structure of papain is also maintained by three , which connect , , and [3]. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures.