Lipase
From Proteopedia
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='1hpl' size='500' side='right' scene=''> |
Revision as of 16:11, 28 March 2012
Lipase is a subclass of esterases which catalyzes the hydrolysis of ester bonds in lipid substrates and is essential for fat digestion. It is primarily produced in the pancreas, but can be found in the mouth and stomach as well. The typically digests fat lipids into monoglycerides and free fatty acids, with the resulting monomers subsequently shuttled to the small intestine and eventually absorbed into the lymphatic system. See also Molecular Playground/Pancreatic Lipase. IntroductionLipase is a hydrolase that catalyzes the breakdown of lipids by hydrolyzing the esters of fatty acids. Lipases are important in digestion, promoting absorption of fats in the intestines. Lipase is primarily found in the pancreas but is also found in the mouth and the stomach. Pancreatic lipase (PDB ID: 1HPL) which is pictured below is a carboxylic ester hydrolase. It is also commonly called pancreatic triacylglycerol lipase and its enzyme class number is E.C. 3.1.1.3 [1]. The reaction catalyzed by this enzyme is shown below. Further breakdown ultimately results in 2-monoacylglycerols and free fatty acids [2]. Pancreatic liapase is a 50 kDa protein, consisting of two identical, 449 residue chains [3]. The determination of the structure and function of lipase was a gradual process. Lipase activity was first demonstrated in the pancreas by Claude Bernard in 1846. It wasn't until 1955 that Mattson and Beck demonstrated a high-specificity of pancreatic lipase for triglyceride primary esters [4]. In recent years, determination of the crystal structure of pancreatic lipase has become the focus and many scientists have worked to further this. Structure
Hydrophobicity/HydrophillicityThe is a useful representation of the distribution of hydrophobic and hydrophillic residues. Hydrophobic residues are shown in red and hydrophillic residues in blue. When the hydrophillic residues are removed and only are shown, it is clear that the core of the enzyme is made of hydrophobic residues while the hydrophillic residues are mainly located on the surface of the enzyme. Lipase Catalytic MechanismAlthough a diverse array of lipase enzymes are found in nature, occupying diverse protein scaffolds, most are built upon an alpha/beta hydrolase fold[5][6] and possess a chymotrypsin-like comprised of an acidic residue, a histidine, and a serine nucleophile. In the case of the images above of a horse pancreatic lipase, the catalytic triad is comprised of [7] This catalytic triad functions like most found in nature, first with the Aspartic acid forming a hydrogen bond with His 263, increasing the pKa of the histidine imidazole nitrogen. This allows the Histidine to act as a powerful general base and deprotonate the serine. The deprotonated serine then can serve as a nucleophile and attack the glycerol backbone of the lipid substrate. A water molecule then donates a proton to the histidine, creating a reactive hydroxyl anion, which can attack the carbonyl carbon of the lipid, releasing the catalytic serine and creating monoglyceride and fatty acid monomers that diffuse away.
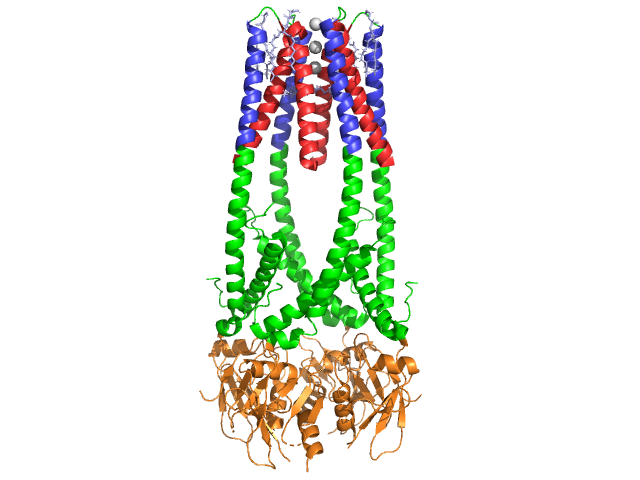
Inhibition of Pancreatic LipaseIn this structure, only one of the two identical chains is shown for lipase and colipase to better visualize the interaction of substrates and ligands with the protein. , a C11 alkyl phosphonate, is a competitive inhibitor of pancreatic lipase which binds to the active site. It is highlighted in purple. There are also five B-octylglucoside molecules in association with lipase. They are shown in grey and red. MUP forms hydrogen bonds with : Ser 152 and His 263, which are part of the catalytic triad, and Phe 77 and Leu 153 which are the stabilizing residues located in the oxyanion hole [8]. added by biochem classReaction 1: Image:M0218.stg01.gif Reaction 2: Reaction 3: Reaction 4: Seen here is the Candida rugosa lipase in with two molecules of cholesteryl linoleate (grey).
Clinical SignificancePancreatic lipase is secreted into the duodenum through the duct system of the pancreas. In a healthy individual, it is in very low concentration in serum. Under extreme disruption of pancreatic function, such as pancreatitis or pancreatic cancer, the pancreas may begin to digest itself and release pancreatic enzymes including pancreatic lipase into serum. Measurement of serum concentration of pancreatic lipase can therefore aid in diagnosis of acute pancreatitis.[9] Here, lipase and colipase can be seen The N-terminal domain also contains a that blocks solvent from entering the active site.
3D Structures of LipaseUpdate November 2011 Eukaryote natives:1hpl – hLip – horse
Prokaryote natives:3guu, 1lbs, 1lbt, 1tca, 1tcb, 1tcc – CaLipA – Candida antarctica Lipase/colipase complexes. The colipase is a co-enzyme whose binding to lipase optimizes the enzymatic activity1n8s – hLip+colipase II Hormone-sensitive-lipases (LIPE) hydrolyze the first fatty acid of the triacylglycerol substrate3k6k – EstE7(LIPE) – metagenome library Putative lipases; Proteins with unknown function but structural similarity to lipase obtained in structural genomics projects.2rau - Lip – Sulfolobus solfataricus Lipase + inhibitors3jwe, 3pe6 - mono-glyceride hLip + SAR629 – covalent inhibitor Lipase conjugated with analogs to its reaction intermediates1lpn, 1lpo, 1lpp – CrLip+ sulfonates Lipase showing bile-salt binding site1aql – cBSSL+taurocholate Lipase with substrate bound at active site2zyh – AfLip (mutant)+fatty acid – Archaeoglobus fulgidus Lipase conjugated to transition-state analogs showing the binding mode of the enzyme catalysis1ys1 – BhLip+hexylphosphonic acid (R) 2-methyl-3-phenylpropyl ester Lipase+lipase chaperone2es4 – Lip+lipase chaperone C-terminal - Burkholderia glumae References
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Quinn R. Murray, Natalie Ziegler, Stephanie Schell, David Canner, Alexander Berchansky, Katelyn Clark, Eric Martz, Leben Tadesse, Joel L. Sussman, Eran Hodis