From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 26: |
Line 26: |
| | | | |
| | ===Catalytic Mechanism=== | | ===Catalytic Mechanism=== |
| - | It was once thought that cysteine proteases, like serine proteases, contained a <scene name='Papain/Active_site/1'>catalytic triad</scene> , consisting of Cys-25, His-159, and Arg-175 <ref> PMID:8140097</ref>. However, site-directed mutagenesis-based studies have demonstrated that Arg-175 is not directly involved in catalysis. Although Arg-175 is clearly important for the enzyme's activity (an Arg175Ala mutation reduces its activity to undetectable levels), this residue neither reacts with the substrate nor modulates the pKa of reacting residues, and therefore cannot be considered catalytic.<ref name="Shokhen">[http://www.ncbi.nlm.nih.gov/pubmed/19688822]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.</ref><ref name="Noble">[http://www.ncbi.nlm.nih.gov/pubmed/11042128]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.</ref> Arg-175 is believed to keep histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual catalytic mechanism.<ref name="U Maine">[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref> Despite this, the basic mechanism of papain-catalyzed proteolysis proceeds much like that of serine proteases. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by an oxyanion hole, formed in large part by <scene name='Papain/Gln_19/1'>Gln-19</scene>. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme. This entire mechanism is shown below<ref name="Harrison">[http://pubs.acs.org/doi/abs/10.1021/ja9711472] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291</ref>.: | + | It was once thought that cysteine proteases, like serine proteases, contained a <scene name='Papain/Catalytic_triad/1'>catalytic triad</scene>, consisting of Cys-25, His-159, and Arg-175 <ref> PMID:8140097</ref>. However, site-directed mutagenesis-based studies have demonstrated that Arg-175 is not directly involved in catalysis. Although Arg-175 is clearly important for the enzyme's activity (an Arg175Ala mutation reduces its activity to undetectable levels), this residue neither reacts with the substrate nor modulates the pKa of reacting residues, and therefore cannot be considered catalytic.<ref name="Shokhen">[http://www.ncbi.nlm.nih.gov/pubmed/19688822]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.</ref><ref name="Noble">[http://www.ncbi.nlm.nih.gov/pubmed/11042128]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.</ref> Arg-175 is believed to keep histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual catalytic mechanism.<ref name="U Maine">[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref> Despite this, the basic mechanism of papain-catalyzed proteolysis proceeds much like that of serine proteases. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by an <scene name='Papain/Oxyanion_hole/1'>oxyanion hole</scene>, formed in large part by Gln-19. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme. This entire mechanism is shown below<ref name="Harrison">[http://pubs.acs.org/doi/abs/10.1021/ja9711472] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291</ref>.: |
| | [[Image:Papainmech6.jpg|400px|center|thumb| General mechanism of papain catalysis <ref name="U Maine" />. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.]] | | [[Image:Papainmech6.jpg|400px|center|thumb| General mechanism of papain catalysis <ref name="U Maine" />. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.]] |
| | | | |
Revision as of 16:09, 6 April 2012
Papain. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings[2]. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, Carica papaya and Vasconcellea cundinamarcensis would have such a practical application beyond proteopedia?
Papain is a 23.4 kDa, 212 residue cysteine protease, also known as papaya proteinase I, from the peptidase C1 family (E.C. 3.4.22.2).[3][4] It is the natural product of the Papaya(Carica papaya)[5], and may be extracted from the plant's latex, leaves and roots. [6]. Papain displays a broad range of functions, acting as an endopeptidase, exopeptidase, amidase, and esterase,[6] with its optimal activity values for pH lying between 6.0 and 7.0, and its optimal temperature for activity is 65 °C. Its pI values are 8.75 and 9.55, and it is best visualized at a wavelength of 278 nm. [5]
Papain's enzymatic use was first discovered in 1873 by G.C. Roy who published his results in the Calcutta Medical Journal in the article, "The Solvent Action of Papaya Juice on Nitrogenous Articles of Food." In 1879, papain was named officially by Wurtz and Bouchut, who managed to partially purify the product from the sap of papaya. It wasn't until the mid-twentieth century that the complete purification and isolation of papain was achieved. In 1968, Drenth et al. determined the structure of papain by x-ray crystallography, making it the second enzyme whose structure was successfully determined by x-ray crystallography. Additionally, papain was the first cysteine protease to have its structure identified.[6] In 1984, Kamphuis et al. determined the geometry of the active site, and the three-dimensional structure was visualized to a 1.65 Angstrom solution.[7] Today, studies continue on the stability of papain, involving changes in environmental conditions as well as testing of inhibitors such as phenylmethanesulfonylfluoride (PMSF), TLCK, TPCK, aplh2-macroglobulin, heavy metals, AEBSF, antipain, cystatin, E-64, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents.[6]
Structure
| General Structural Features
Papain is a relatively simple enzyme, consisting of a single 212 residue chain. A majority of papain's residues are as shown in purple. As with all proteins, it is primarily the exclusion of these residues by water that leads to papain's assumption of a globular form. Despite its apparent simplicity and small size, papain folds into two distinct, evenly sized , each with its own (surface residues are transparent, hydrophobic-core residues are colored and opaque, and the remaining are polar, non-surface residues).[7]
These two subunits are held together with , where each protein domain holds the opposite domain. In papain's case the "arm" crossing primarily occurs on or near the surface.[8] It is between these two domains that the is situated. The two domains interact with one another via hydrophobic interactions, (shown in white), and electrostatic interactions in this cleft. For example, from the L Domain hydrophobically interacts with the carbon atoms on residues Lys174, Ala162 and Pro129 of the R Domain. hydrogen bonds multiple times with the oxygen atoms of Ser176 and also with the oxygen atom on Tyr88. Electrostatic interactions are seen between where the carboxyl group of Glu35 forms an ionic bond with the ammonia group of the Lys174 residue. The of interactions within the cleft between the two domains ensures that the lobes do not move with respect to one another. [9]
In addition to hydrophobic residues, papain contains a variety of , some carrying a , shown in gray at physiological pH, and are therefore acidic; others a , shown in purple, and are therefore basic. The rest of the , shown in a light gray, are neutral. As expected, the charged face outward due to their hydrophilic nature.
Papain's secondary structure is composed of 21% (45 residues comprising 17 sheets) and 25% (51 residues comprising 7 helices). The rest of the residues, accounting for over 50% of the enzymes structure, make up ordered non-repetative sequences.[10] These secondary structures may be traced from the N- to C-terminus by means of . As shown in this scene, the red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the blue end at the C-terminus. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the (residues 25-42) is maintained as a result of between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many (red residues are hydrophilic).
strongly contribute to the tertiary structure of papain.[5] In this particular image, clarification of residue coordination is demonstrated by color: paired residues are shown in the same color, oxygen is shown in red, and nitrogen is shown in blue. The tertiary structure of papain is also maintained by three , which connect , , and [4]. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures.
Substrate Binding
Papain has a relatively small consisting of three residues: Cys-25, His-159, and Arg-175. The sulfhydryl group on Cys-25 often forms covalent bonds with substrates. His-159 supports Cys-25, and while Arg-175 does not directly participate in the catalytic mechanism, it keeps histidine-159 in its stabilized imidazole form. In addition to the active site, sometimes referred to as the catalytic site, Papain consists of many Subsites are defined as the regions on the enzyme surface which interact with one amino acid residue of the substrate. In the case of Papain, there are seven subsites labeled as follows: S1, S2, S3, S4, located on the amino side of the catalytic site; S1', S2', and S3', located on the carboxyl side of the catalytic site. The binding sites of a substrate are labeled according to how they fit into the binding cleft. P1 associates with S1, P2 with S2, etc. All seven of Papain's subsites hydrogen bond to the corresponding substrate P subsites. The following of Papain have been identified as follows: S1 - His-159; S2 - Trp-177; S3 - Gln-19; S4 - Gly-23; S2' - Asp-158; and S3' - Asp-64. Unfortunately, S1' has yet to be identified.
The seven subsites of Papain have various preferences for substrate residues. Through a variety of experiments, Berger & Schechter[11], were able to show that S1 binds alanine better than glycine, and the larger side chains of lysine, arginine, leucine, and phenylalanine better than alanine. Thus showing that binding in S1 is predominantly hydrophobic. S2 prefers a phenylalanine or a valine residue. Interestingly enough, S2 binds to hydrophobic residues of both short and long peptide chains. They were able to show that subsites S1' and S2' are strongly stereospecific. The conclusion of their research was that Papain's binding site residues show a strong stereospecificity, special interactions, and space limitations.[12]
Catalytic Mechanism
It was once thought that cysteine proteases, like serine proteases, contained a , consisting of Cys-25, His-159, and Arg-175 [13]. However, site-directed mutagenesis-based studies have demonstrated that Arg-175 is not directly involved in catalysis. Although Arg-175 is clearly important for the enzyme's activity (an Arg175Ala mutation reduces its activity to undetectable levels), this residue neither reacts with the substrate nor modulates the pKa of reacting residues, and therefore cannot be considered catalytic.[14][15] Arg-175 is believed to keep histidine-159 in its stabilized imidazole form, while both histidine-159 and cysteine-25 take part in the actual catalytic mechanism.[16] Despite this, the basic mechanism of papain-catalyzed proteolysis proceeds much like that of serine proteases. The mechanism begins when a peptide binds to the active site. Cys-25 is then deprotonated by His-159 and attacks the substrate carbonyl carbon. This forms a covalent, tetrahedral intermediate that is stabilized by an , formed in large part by Gln-19. Next, His-159 acts as a general acid, protonating the nitrogen in the peptide bond, which acts as a leaving group as the carbonyl reforms. This now free C-terminal portion of the peptide is released. Water then enters the active site and attacks the carbonyl carbon while it is deprotonated by His-159, again forming an oxyanion hole-stabilized tetradral covalent intermediate. Finally, the carbonyl reforms and the Cys-25 sulfur acts as a leaving group, releasing the N-terminal portion of the peptide and regenerating the enzyme. This entire mechanism is shown below[17].:
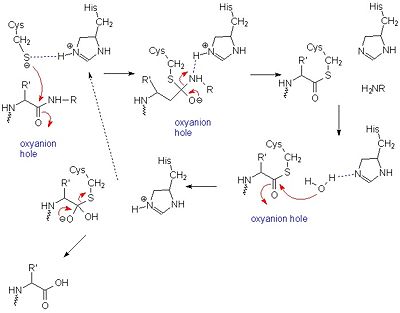 General mechanism of papain catalysis [16]. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.
|
Inhibitors
Leupeptin
is a commonly studied broad-spectrum competitive protease inhibitor first crystallized by Schröder et. al. It inhibits by binding and interacting with the active site which allows it to block the enzyme's desired protein substrate from binding. There are many that interact with leupeptin, which predominantly interact (interactions in blue), including tyrosine, tryptophan, and valine, which coordinate the bound leuptin. Some of the enzyme's residues also make . These hydrogen bonds, shown in white, include interactions between hydrogens on both Gln-19 and the amide nitrogen of the catalytic Cys-25 with the arginal carbanion, forming the catalytically important oxyanion hole. In addition, Gly-66 interacts with the second leucine in leupeptin while Asp-158 interacts with a hydrogen on the arginal. These interactions further stabilize and orient the substrate in the binding pocket[18].
A recent study has shown that Leupeptin forms a covalent bond between its and the hydrogen in Cys-25. The inhibitor has the structure Ac-Leu-Leu-Arginal, where Ac is an acetyl group attached to the nitrogen of the first leucine. Cysteine-25, which acts as a catalytic nucleophile, attacks the arginal aldehyde forming a tight-binding transition state from which the normal catalytic mechanism cannot proceed due to this carbonyl having no potential leaving groups bonded to it.
Cathepsin K
The goal of research for the development of an inhibitor for is the hope to develop a treatment for osteoporosis. In two different Cathepsin K inhibitors, , 1BP4, and , 1BQI, it is evident that the inhibitor binds with much closer proximity than that of Clik148. 1BP4, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, inhibits by interacting with 11 different residues on papain: Gln19, Gly20, Ser21, Gly23, Asn64, Gly65, Gln142, Asp158, His159, Trp177, and Trp181. These interactions range from hydrophobic, electrostatic, and hydrogen bonding, to , illustrated in blue, between the aromatic ring of the carbobenzyl group on 1BP4, and Trp-177 of papain. The inhibition of papain by IBQI, carbobenzyloxy-(L)-leucinyl-(L)leucinyl methoxymethylketone, is quite similar to that of IBP4, although it does not bind quite as tightly. It binds to seven residues of papain: Gln19, Gly23, Gly65, Gln142, His159, Trp177, Trp181. Additionally, it has similar , shown in blue, between the Cbz ring on the inhibitor and Trp 177, though it is more difficult to visualize with the given PDB file.[19]
Cathepsin L
Cathepsin L is another inhibitor of the papain enzyme. interacts with the Gln19, Cys25, Gly66, Asp158, and Trp177 by hydrogen bonding them (Cathepsin L is illustrated in CPK coloring while the interacting sites of Papain are also shown in CPK. In addition to hydrogen bonding, hydrophobic interactions exist to exclude water, allowing the papain enzyme and Cathepsin L to associate even closer. Finally, between Trp177 and the Capthespin L molecule hold them tightly together.[20] Cathepsin L plays a roll in many different diseases including malaria, leishmaniasis, Chagas' disease, African trypanosomiasis, toxoplasmosis, and amoebiasis. Some studies show that there is a relation between cathepsins and certain cancers, alzheimer's, and arthritis. [21]
Stefin B
acts as a competitive inhibitor to cysteine proteases-- it binds tightly but reversibly to the papain active site. Stefin inhibitors are characterized by Mr of about 11,000, no disulfide bonds and no associated carbohydrates.
In Stefin B, the Gly9 residue along with form a "wedge" complementary to the active site groove of papain. This wedge makes extensive and tight interactions with papain and a total of 128 intermolecular atom-atom interactions occur. Met6-Pro11, Gln53-Asn59, Gln101-His104 and Tyr124-Phe125 on the wedge all have some interaction to the enzyme though not always direct. All residues from the base and both sides of the are involved in the complex with the inhibtor (Trp177, Ser21, Cys63, Cys25, Asp158 and His159).
There are a small number of between stefin B and papain, however there are many more polar interactions mediated by . Thirteen solvent molecules bridge polar residues of the enzyme and inhibitor. Seventeen hydrogen bonds are made with a solvent molecule and stefin. Fourteen of these bridges form a papain contact. The rest of the interactions are largely hydrophobic-- involving apolar . [22]
| |
Common Uses
Medicinal
Papain has been used for a plethora of medicinal purposes including treating inflammation, shingles, diarrhea, psoriasis, parasites, and many others.
[23] One major use is the treatment of cutaneous ulcers including diabetic ulcers and pressure ulcers. Pressures ulcers plague many bed bound individuals and are a major source of pain and discomfort. Two papain based topical drugs are Accuzyme and Panafil, which can be used to treat wounds like cutaneous ulcers.
[25]
A recent New York Times article featured papain and other digestive enzymes. With the number of individuals suffering from irritable bowel syndrome and other gastrointestinal issues, many people are turning toward natural digestive aid supplements like papain. The author even talks about the use of papain along with a pineapple enzyme, bromelain, in cosmetic facial masks. Dr. Adam R. Kolker (a plastic surgeon) is quoted in the article saying that "For skin that is sensitive, enzymes are wonderful." He bases these claims off the idea that proteases like papain help to break peptide bonds holding dead skin cells to the live skin cells.[26]
Commercial and Biomedical
Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains. This non-specific nature of papain's hydrolase activity has led to its use in many and varied commercial products. It is often used as a meat tenderizer because it can hydrolyze the peptide bonds of collagen, elastin, and actomyosin. It is also used in contact lens solution to remove protein deposits on the lenses and marketed as a digestive supplement. [23]
Despite a low percentage of sequence identities, inhibition and sequence analyses have increasingly been drawing parallels between L proteinases, that involve the foot-and-mouth disease virus and equine rhinovirus 1, and papain. With a similar overall fold to papain and identifiable regions that resemble papain's five alpha-helices and seven beta-sheets, L proteinases of foot-and-mouth disease virus and of equine rhinovirus 1 reveal a mode of operation that is very papain-like. [27]
References
- ↑ [1] Papaya's Nutrition Facts
- ↑ [2] Ameridan International
- ↑ [3] Uniprot
- ↑ 4.0 4.1 [4] 9PAP PDB
- ↑ 5.0 5.1 5.2 [5] Sigma Aldrich
- ↑ 6.0 6.1 6.2 6.3 [6] Worthington
- ↑ 7.0 7.1 Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233-56. PMID:6502713
- ↑ [7] Jane S. Richardson
- ↑ [8] The Structure of Papain
- ↑ [9] RCSB PDB
- ↑ Berger A, Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249-64. PMID:4399049
- ↑ Berger A, Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249-64. PMID:4399049
- ↑ Wang J, Xiang YF, Lim C. The double catalytic triad, Cys25-His159-Asp158 and Cys25-His159-Asn175, in papain catalysis: role of Asp158 and Asn175. Protein Eng. 1994 Jan;7(1):75-82. PMID:8140097
- ↑ [10]Shokhen M, N Khazanov, and A Albeck. 2009. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 77(4):916-26.
- ↑ [11]Noble MA, Gul S, Verma CS, Brocklehurst K. 2000. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000 351: 723-33.
- ↑ 16.0 16.1 [12] University of Maine
- ↑ [13] Harrison, M.J., N.A. Burton, and I.H. Hillier. 1997. Catalytic Mechanism of the Enzyme Papain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J. Am. Chem. Soc. 119: 12285-12291
- ↑ [14] Schröder, E., C. Phillips, E. Garman, K. Harlos, C. Crawford. 1997. X-ray crystallographic structure of a papain-leupeptin complex. FEBS Letters 315: 38-42
- ↑ LaLonde JM, Zhao B, Smith WW, Janson CA, DesJarlais RL, Tomaszek TA, Carr TJ, Thompson SK, Oh HJ, Yamashita DS, Veber DF, Abdel-Meguid SS. Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites. J Med Chem. 1998 Nov 5;41(23):4567-76. PMID:9804696 doi:10.1021/jm980249f
- ↑ Beavers MP, Myers MC, Shah PP, Purvis JE, Diamond SL, Cooperman BS, Huryn DM, Smith AB 3rd. Molecular docking of cathepsin L inhibitors in the binding site of papain. J Chem Inf Model. 2008 Jul;48(7):1464-72. Epub 2008 Jul 4. PMID:18598021 doi:10.1021/ci800085c
- ↑ Valadares NF, Dellamano M, Soares-Costa A, Henrique-Silva F, Garratt RC. Molecular determinants of improved cathepsin B inhibition by new cystatins obtained by DNA shuffling. BMC Struct Biol. 2010 Sep 30;10:30. PMID:20920298 doi:10.1186/1472-6807-10-30
- ↑ Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990 Jun;9(6):1939-47. PMID:2347312
- ↑ 23.0 23.1 [15] Web MD
- ↑ [16] RX
- ↑ [17] National PBM Drug Monograph
- ↑ [18] Enzymes Try to Grab the Spotlight
- ↑ Skern T, Fita I, Guarne A. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J Gen Virol. 1998 Feb;79 ( Pt 2):301-7. PMID:9472614
3D Structures of Papain
3LFY, 1KHP, 1KHQ, 1CVZ, 1BQI, 1BP4, 1PPN, 1PPP, 1PIP, 1POP, 1PE6, 9PAP, 1PPD, 1PAD, 2PAD, 4PAD, 5PAD, 6PAD- Carica papaya
3IMA - Colocasia esculenta
1STF - Homo sapiens
2CIO - Trypanosoma brucei
3E1Z - Trypanosoma cruzi
Page seeded by OCA on Tue Feb 17 04:20:31 2009