User:Jamie Abbott/Sandbox2
From Proteopedia
| Line 1: | Line 1: | ||
== Histidyl-tRNA Synthetase == | == Histidyl-tRNA Synthetase == | ||
| - | Histidyl tRNA Synthetase (HisRS) is a 94kD <scene name='User:Jamie_Abbott/Sandbox2/Hisrsdimer/2'>homodimer</scene> that belongs to the class II of aminoacyl-tRNA synthetases (aaRS). [http://www.pdb.org/pdb/101/motm.do?momID=16 Aminoacyl-tRNA synthetases] Aminoacyl-tRNA synthetases have been partitioned into two classes, containing 10 members, on the basis of sequence comparisons<ref name="Eriani">PMID: 2203971</ref>. Class I and Class II differ mainly with respect to the topology of the catalytic fold and site of esterification on cognate tRNA<ref name="Eriani" />. Class II enzymes have a <scene name='User:Jamie_Abbott/Sandbox2/Catalytic_domain/1'>catalytic domain</scene> composed of anti-parallel β-sheets and α-helices (residues 1-325). Additionally, class II enzymes can be further divided into three subgroups: class IIa, distinguished by an N-terminal catalytic domain and C-terminal accessory domain (later shown to be anticodon binding domain); class IIb, whose anticodon binding domain is located on the N-terminal side of the fold; and class IIc, encompassing the tetrameric PheRS and GlyRS class II synthetases.<ref name="Cusack91">PMID: 1852601</ref> | + | '''Histidyl tRNA Synthetase (HisRS)''' is a 94kD <scene name='User:Jamie_Abbott/Sandbox2/Hisrsdimer/2'>homodimer</scene> that belongs to the class II of aminoacyl-tRNA synthetases (aaRS). [http://www.pdb.org/pdb/101/motm.do?momID=16 Aminoacyl-tRNA synthetases] Aminoacyl-tRNA synthetases have been partitioned into two classes, containing 10 members, on the basis of sequence comparisons<ref name="Eriani">PMID: 2203971</ref>. Class I and Class II differ mainly with respect to the topology of the catalytic fold and site of esterification on cognate tRNA<ref name="Eriani" />. Class II enzymes have a <scene name='User:Jamie_Abbott/Sandbox2/Catalytic_domain/1'>catalytic domain</scene> composed of anti-parallel β-sheets and α-helices (residues 1-325). Additionally, class II enzymes can be further divided into three subgroups: class IIa, distinguished by an N-terminal catalytic domain and C-terminal accessory domain (later shown to be anticodon binding domain); class IIb, whose anticodon binding domain is located on the N-terminal side of the fold; and class IIc, encompassing the tetrameric PheRS and GlyRS class II synthetases.<ref name="Cusack91">PMID: 1852601</ref> |
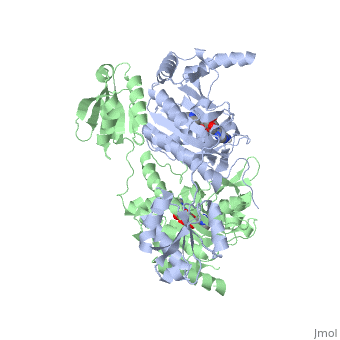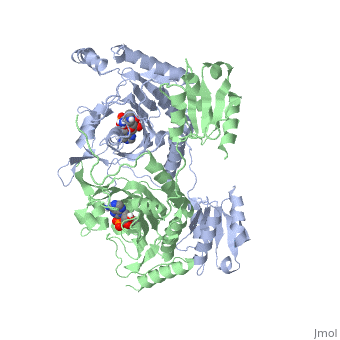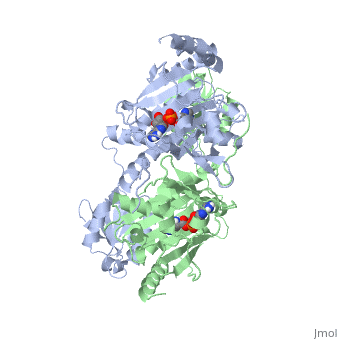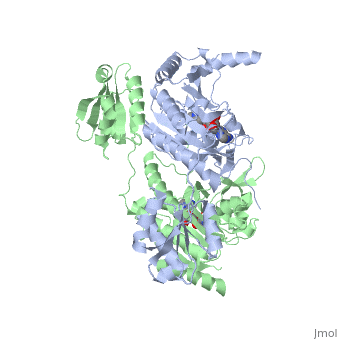
<StructureSection load='1KMM' size='500' side='right' caption='Structure of Histidyl-tRNA Synthetase (PDB entry [[1KMM]])' scene=''>Class II aminoacyl-tRNA synthetases aminoacylate the 3'OH of their cognate tRNAs. | <StructureSection load='1KMM' size='500' side='right' caption='Structure of Histidyl-tRNA Synthetase (PDB entry [[1KMM]])' scene=''>Class II aminoacyl-tRNA synthetases aminoacylate the 3'OH of their cognate tRNAs. | ||
Revision as of 16:54, 15 April 2012
Contents |
Histidyl-tRNA Synthetase
Histidyl tRNA Synthetase (HisRS) is a 94kD that belongs to the class II of aminoacyl-tRNA synthetases (aaRS). Aminoacyl-tRNA synthetases Aminoacyl-tRNA synthetases have been partitioned into two classes, containing 10 members, on the basis of sequence comparisons[1]. Class I and Class II differ mainly with respect to the topology of the catalytic fold and site of esterification on cognate tRNA[1]. Class II enzymes have a composed of anti-parallel β-sheets and α-helices (residues 1-325). Additionally, class II enzymes can be further divided into three subgroups: class IIa, distinguished by an N-terminal catalytic domain and C-terminal accessory domain (later shown to be anticodon binding domain); class IIb, whose anticodon binding domain is located on the N-terminal side of the fold; and class IIc, encompassing the tetrameric PheRS and GlyRS class II synthetases.[2]
| |||||||||||
Mechanism
Electrophilic Catalysis
The HisRS active site contains a highly conserved residue, Arg259, takes part in electrophilic catalysis for the adenylation reaction. First, as Arg259 is positioned on the HisA loop serves to fix the α-carboxylate group of the histidine substrate as the attacking nucleophile[6]. Second, the guanidinium group of Arg259 is positioned approximately 3 Å from the α-phosphate of ATP where it serves as the electrophilic catalyst. Arg113 as well as Arg259 are arranged to interact with α-phosphate of ATP and thereby stabilize negative charge developed on the non-bridging oxygens during the transition state aarsbk. Evidence for Arg259 playing a critical role in catalysis is observed in a two or three log decrease in activity when substituted with a histidine [4] or other amino acids[7]. Arg259 also interacts with the phenolic OH of Tyr264, which in turn donates a hydrogen bond to the Nδ of the histidine substrate[3]. Utilizing Arg259 for catalysis is unique to HisRS as other class II aaRS enzymes, AspRS[8] and SerRS[9], which use a divalent magnesium metal ion to coordinate the α-phosphate of ATP and serve as an electrophilic catalysis.
Evolutionary Conservation
Structural Homology
3D Structures of Histidyl-tRNA Synthetase
Bacteria
Eukaryota
Archara
References
- ↑ 1.0 1.1 Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203-6. PMID:2203971 doi:http://dx.doi.org/10.1038/347203a0
- ↑ Cusack S, Hartlein M, Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489-98. PMID:1852601
- ↑ 3.0 3.1 3.2 3.3 Francklyn, C., and Arnez, J.G. (2004) in Aminoacyl-tRNA Synthetases (Ibba, M.,Francklyn, C.,Cusack, S.. Eds.) Landes Publishing, Austin, TX
- ↑ 4.0 4.1 Arnez JG, Augustine JG, Moras D, Francklyn CS. The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1997 Jul 8;94(14):7144-9. PMID:9207058
- ↑ Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997 Jun;22(6):211-6. PMID:9204708
- ↑ Arnez JG, Flanagan K, Moras D, Simonson T. Engineering an Mg2+ site to replace a structurally conserved arginine in the catalytic center of histidyl-tRNA synthetase by computer experiments. Proteins. 1998 Aug 15;32(3):362-80. PMID:9715912
- ↑ Ruhlmann A, Cramer F, Englisch U. Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli. Biochem Biophys Res Commun. 1997 Aug 8;237(1):192-201. PMID:9266856 doi:10.1006/bbrc.1997.7108
- ↑ Poterszman A, Delarue M, Thierry JC, Moras D. Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J Mol Biol. 1994 Nov 25;244(2):158-67. PMID:7966328 doi:http://dx.doi.org/10.1006/jmbi.1994.1716
- ↑ Belrhali H, Yaremchuk A, Tukalo M, Berthet-Colominas C, Rasmussen B, Bosecke P, Diat O, Cusack S. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure. 1995 Apr 15;3(4):341-52. PMID:7613865