DOPA decarboxylase
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
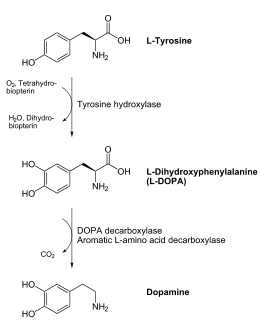
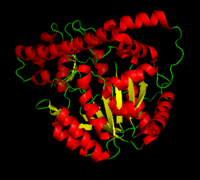
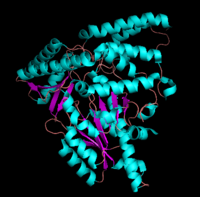
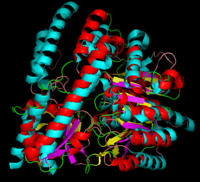
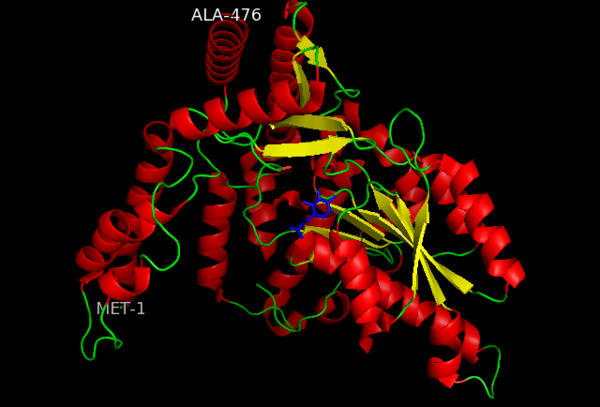
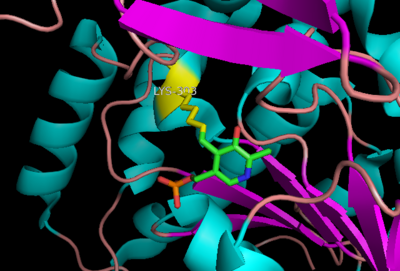
[[image:dopa.png|thumb|left|300px|'''Dopamine Synthesis''']]'''DOPA decarboxylase''' (DDC, aromatic L-amino acid decarboxylase, tryptophan decarboxylase, 5-hydroxytryptophan decarboxylase, AAAD) ([[EC Number|EC]] 4.1.1.28) is an approximately 104 kDa protein that belongs to the '''aspartate aminotransferase family''' (fold type 1) of '''[http://en.wikipedia.org/wiki/Pyridoxal_phosphate ''PLP'']-dependent''' (vitamin B6-dependent) enzymes. The catalytically active form of the enzyme exists as a homodimer, typical of this class of enzymes.<ref name="schneider">PMID:10673430 </ref> The homodimeric form of the enzyme purified from [http://en.wikipedia.org/wiki/Sus_scrofa ''sus scrofa''] is shown in complex with the inhibitor '''[http://en.wikipedia.org/wiki/Carbidopa ''carbidopa'']''' to the right. | [[image:dopa.png|thumb|left|300px|'''Dopamine Synthesis''']]'''DOPA decarboxylase''' (DDC, aromatic L-amino acid decarboxylase, tryptophan decarboxylase, 5-hydroxytryptophan decarboxylase, AAAD) ([[EC Number|EC]] 4.1.1.28) is an approximately 104 kDa protein that belongs to the '''aspartate aminotransferase family''' (fold type 1) of '''[http://en.wikipedia.org/wiki/Pyridoxal_phosphate ''PLP'']-dependent''' (vitamin B6-dependent) enzymes. The catalytically active form of the enzyme exists as a homodimer, typical of this class of enzymes.<ref name="schneider">PMID:10673430 </ref> The homodimeric form of the enzyme purified from [http://en.wikipedia.org/wiki/Sus_scrofa ''sus scrofa''] is shown in complex with the inhibitor '''[http://en.wikipedia.org/wiki/Carbidopa ''carbidopa'']''' to the right. | ||
| - | DOPA decarboxylase is responsible for the synthesis of '''[http://en.wikipedia.org/wiki/Dopamine ''dopamine'']''' and [http://en.wikipedia.org/wiki/Serotoninn ''serotonin''] from '''[http://en.wikipedia.org/wiki/L-dopa ''L-DOPA'']''' and [http://en.wikipedia.org/wiki/L-5-Hydroxytryptophan ''L-5-hydroxytryptophan''], respectively. | + | DDC catalyzes the conversion of aromatic amino acids into their corresponding amines. DOPA decarboxylase is responsible for the synthesis of '''[http://en.wikipedia.org/wiki/Dopamine ''dopamine'']''' and [http://en.wikipedia.org/wiki/Serotoninn ''serotonin''] from '''[http://en.wikipedia.org/wiki/L-dopa ''L-DOPA'']''' and [http://en.wikipedia.org/wiki/L-5-Hydroxytryptophan ''L-5-hydroxytryptophan''], respectively. It is highly stereospecific, yet relatively nonspecific in terms of substrate, making it a somewhat uninteresting enzyme to study. Although it is not typically a rate-determining step of dopamine synthesis, the decarboxylation of L-DOPA to dopamine by DDC is the controlling step for individuals with '''[http://en.wikipedia.org/wiki/Parkinson%27s_disease ''Parkinson's disease'']'''<ref name="hadjiiconstantinou">PMID:1904055 </ref> , the second most common neurodegenerative disorder, occuring in 1% of the population over the age of 65. The loss of dopaminergic neurons is the main cause of cognitive impairment and tremors observed in patients with the disease. The hallmark of the disease is the formation of [http://en.wikipedia.org/wiki/alpha-synuclein ''alpha-synuclein''] containing [http://en.wikipedia.org/wiki/Lewy bodies ''Lewy bodies'']. |
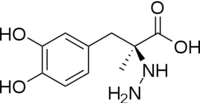
| + | Currently, treatment for the disease is aimed at DOPA decarboxylase inhibition. Since dopamine cannot cross the blood-brain barrier, it cannot be used to directly treat Parkinson's disease. Thus, exogenously administered L-DOPA is the primary treatment for patients suffering from this neurodegenerative disease. Unfortunately, DOPA decarboxylase rapidly converts L-DOPA to dopamine in the blood stream, with only a small percentage reaching the brain. By inhibiting the enzyme, greater amounts of exogenously administered L-DOPA can reach the brain, where it can then be converted to dopamine. <ref name="burkhard">PMID:11685243 </ref> . See also [[DOPA Decarboxylase]] and [[User:Brian Hernandez/DOPA Decarboxylase]]. Unfortunately, with continued L-Dopa treatment, up to 80% of patients experience 'wearing-off' symptoms, dyskinesias and other motor complications (referred to as the "on-off phenomenon". <ref name="lees">PMID:1904055 </ref>. Clearly, a better understanding of the catalytic mechanism and enzymatic activity of DDC in both healthy and PD individuals is critical to drug design and treatment of the disease. | ||
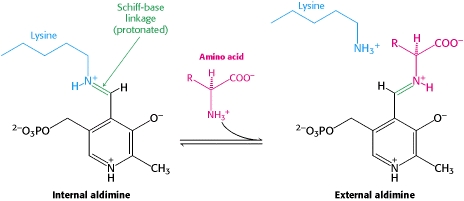
==PLP-Dependent Enzymes== | ==PLP-Dependent Enzymes== | ||
Revision as of 15:21, 30 April 2012
| |||||||||||
3D structures of DOPA decarboxylase
Update November 2011
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
- ↑ 8.0 8.1 Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez