We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 497
From Proteopedia
(Difference between revisions)
(→References) |
(→References) |
||
| Line 25: | Line 25: | ||
<references/> | <references/> | ||
| - | |||
| - | Image from the RCSB PDB (www.pdb.org) of PDB ID 3TFH (Schuller, D.J., Reisch, C.R., Moran, M.A., Whitman, W.B., Lanzilotta, W.N. (2012) Structures of dimethylsulfoniopropinate-dependent demethylase from the marine organism pelagabacter ubique. Protein Sci. 21: 289-298). | ||
| - | |||
| - | Schuller, D.J., Reisch, C.R., Moran, M.A., Whitman, W.B., Lanzilotta, W.N. (2012) Structures of dimethylsulfoniopropinate-dependent demethylase from the marine organism pelagabacter ubique. Protein Sci. 21: 289-298. | ||
Revision as of 15:19, 1 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Dimethylsulfoniopropionate-Dependent Demethylase A (DmdA))
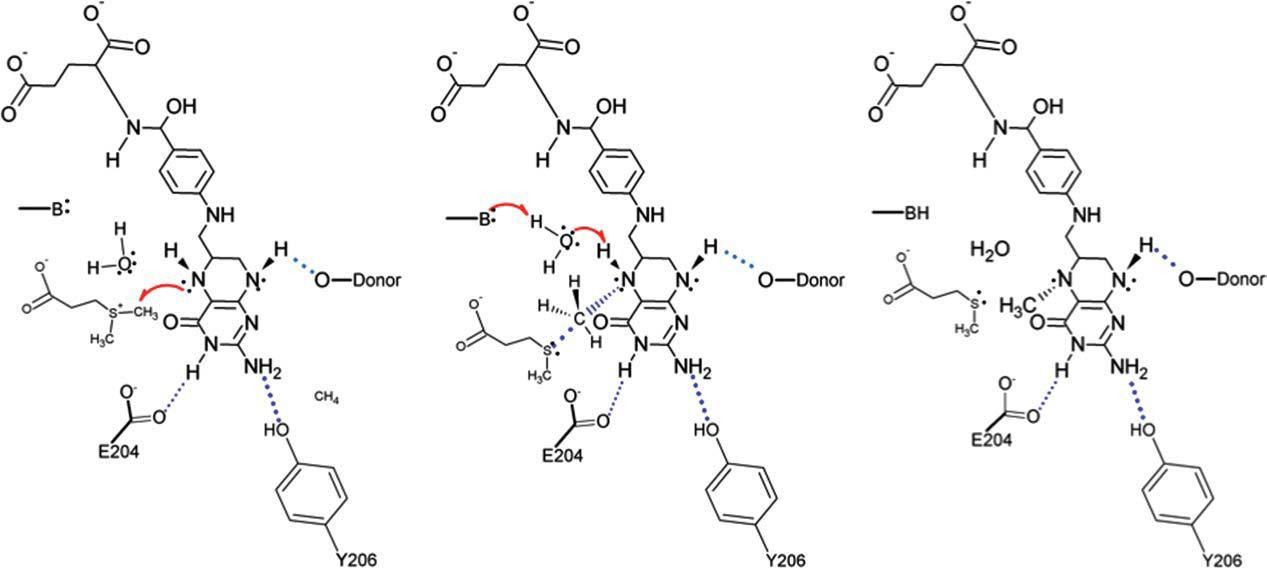
IntroductionDimethylsulfoniopropionate-Dependendent Demethylase A (DmdA) is an enzyme produced by marine bacterioplankton in order to demethylate Dimethylsulfoniopropionate (DMSP). StructureThe structure of DmdA was recently solved through X-Ray Diffraction [1]. The structure contains 369 amino acids folded into four domains and containing two ligands. Mechanism of ActionThe specific mechanism of DmdA is still being investigated. However, a mechanism was recently proposed [2] Possible ApplicationsReferences
|