Sandbox Reserved 497
From Proteopedia
| Line 12: | Line 12: | ||
==Structure== | ==Structure== | ||
| - | The structure of DmdA has recently been solved through the use of X-Ray diffraction | + | The structure of DmdA has recently been solved through the use of X-Ray diffraction <ref> Image from the RCSB PDB (www.pdb.org) of PDB ID 3TFH (Schuller, D.J., Reisch, C.R., Moran, M.A., Whitman, W.B., Lanzilotta, W.N. (2012) Structures of dimethylsulfoniopropinate-dependent demethylase from the marine organism pelagabacter ubique. Protein Sci. 21: 289-298). </ref>. The structure is composed of 369 amino acid residues and contains three distinct domains and four ligands, two of which are sodium ions and two of which are glycerol. While DmdA belongs to the glycine cleavage T-protein (GcvT) family there is only approximately 25% sequence identify. These few conserved amino acids likely interact with tetrahydrofolate (THF), which is a cofactor required by DmdA as well as many other enzymes in the GcvT family. |
==Mechanism of Action== | ==Mechanism of Action== | ||
Revision as of 07:34, 2 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Dimethylsulfoniopropionate-Dependent Demethylase (DmdA)
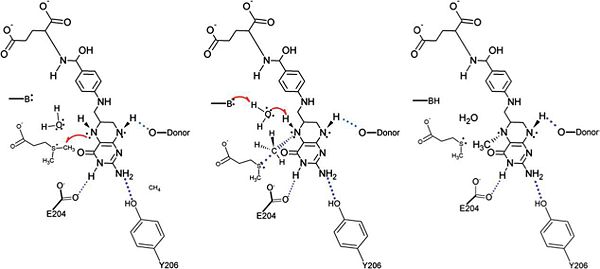
IntroductionDimethylsulfoniproprionate (DMSP) is a common metabolite produced by marine microorganisms and it acts as a significant carbon and sulfur source for marine bacteria. Degradation of DMSP occurs by either the cleavage pathway or the demethylation pathway [1]. The demethylation pathway is characterized by the conversion of DMSP into methylmercaptopropionate (MMPA). Dimethylsulfoniopropionate-Dependendent Demethylase (DmdA) is the first enzyme in the demethylation pathway and facilitates this conversion by acting as a methyl transferase. StructureThe structure of DmdA has recently been solved through the use of X-Ray diffraction [2]. The structure is composed of 369 amino acid residues and contains three distinct domains and four ligands, two of which are sodium ions and two of which are glycerol. While DmdA belongs to the glycine cleavage T-protein (GcvT) family there is only approximately 25% sequence identify. These few conserved amino acids likely interact with tetrahydrofolate (THF), which is a cofactor required by DmdA as well as many other enzymes in the GcvT family. Mechanism of ActionThe specific mechanism of DmdA is still being investigated. However, a mechanism was recently proposed [3] Possible ApplicationsReferences
|