Sandbox Reserved 494
From Proteopedia
| Line 9: | Line 9: | ||
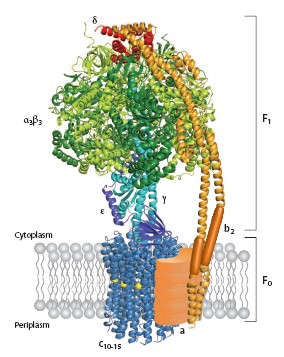
Strucutre of ATP synthases are basically similiar whatever the source. In their simplest form in prokaryotes, they contain eight different subunits, with stoichiometry α<sub>3</sub>β<sub>3</sub>γδεab<sub>2</sub>c<sub>10-15</sub>. The total molecular size is about 530kDa<ref name="MM">PMID: 11997128</ref>. The enzyme consists of the extramembranous F<sub>1</sub> catalytic domain linked by means of a central stalk to an intrinsic membrane domain called F<sub>0</sub>. In the atomic structure of F<sub>1</sub>, the α and β subunits are arranged alternately around a coiled coil of two antiparallel α helices in the γ subunit <ref name="CV" PMID: 19489730</ref>.The catalytic sites are in the β subunits at the α/β subunit interface. The remainder of the γ subunit protrudes from the α<sub>3</sub>β<sub>3</sub> assembly and can be cross linked to the polar loop region of the c subunits in F<sub>0</sub>. In mitochondria, the δ and ε subunits are associated with the γ subunit in the central stalk assembly, as are the bacterial and chloroplast ε subunits, the counterparts of mitochondrial δ. ATP-dependent rotation of γ and ε within an immobilized α<sub>3</sub>β<sub>3</sub> complex from the thermophilic bacterium '''<FONT COLOR="#F535AA">''Bacillus'' PS3</FONT>''' has been observed directly. | Strucutre of ATP synthases are basically similiar whatever the source. In their simplest form in prokaryotes, they contain eight different subunits, with stoichiometry α<sub>3</sub>β<sub>3</sub>γδεab<sub>2</sub>c<sub>10-15</sub>. The total molecular size is about 530kDa<ref name="MM">PMID: 11997128</ref>. The enzyme consists of the extramembranous F<sub>1</sub> catalytic domain linked by means of a central stalk to an intrinsic membrane domain called F<sub>0</sub>. In the atomic structure of F<sub>1</sub>, the α and β subunits are arranged alternately around a coiled coil of two antiparallel α helices in the γ subunit <ref name="CV" PMID: 19489730</ref>.The catalytic sites are in the β subunits at the α/β subunit interface. The remainder of the γ subunit protrudes from the α<sub>3</sub>β<sub>3</sub> assembly and can be cross linked to the polar loop region of the c subunits in F<sub>0</sub>. In mitochondria, the δ and ε subunits are associated with the γ subunit in the central stalk assembly, as are the bacterial and chloroplast ε subunits, the counterparts of mitochondrial δ. ATP-dependent rotation of γ and ε within an immobilized α<sub>3</sub>β<sub>3</sub> complex from the thermophilic bacterium '''<FONT COLOR="#F535AA">''Bacillus'' PS3</FONT>''' has been observed directly. | ||
| - | [[Image:1c17_dimor_(3).jpg | thumb|frame|Two structural domains of ATP synthase | + | [[Image:1c17_dimor_(3).jpg | thumb|frame|Two structural domains of ATP synthase.]] |
==High resolution structure analysis== | ==High resolution structure analysis== | ||
Revision as of 17:22, 2 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
>
BackgroundATP synthesis is the most prevalent chemical reaction in the biological world and ATP synthase is one of the most ubiquitous, abuntant proteins on earth. From Escherichia coli to plants and mammals, this enzyme is one of the most conserved during evolution[1].PDB codes are 1c17. The molecular study of ATP synthase was initiated in 1960 when Efraim Racker and his colleagues reported thia isolation of soluable factor from beef heart mitochondria. ATP synthase produces ATP from adenosine diphosphate (ADP) and inorganic phosphate with the use of energy from a transmembrane proton-motive force generated by respiration or photosynthase[2]. Structure of ATP synthaseStrucutre of ATP synthases are basically similiar whatever the source. In their simplest form in prokaryotes, they contain eight different subunits, with stoichiometry α3β3γδεab2c10-15. The total molecular size is about 530kDa[3]. The enzyme consists of the extramembranous F1 catalytic domain linked by means of a central stalk to an intrinsic membrane domain called F0. In the atomic structure of F1, the α and β subunits are arranged alternately around a coiled coil of two antiparallel α helices in the γ subunit [4]. PerspectiveAn emerging possibility of step-size mismatch between the F1 and F0 motors provides an opportunity to find a noval coupling mechanism of the two motors that will explain why the mismatch is good for the enzyme. Finally, one can even dream of using this, the world's tiniest motor, as an engine part in the fabrication of nano-machines. The marvel of ATP will continue. Additional ResourcesFor additional information, see: Photosynthesis
References
[1] ATP synthase-a marvellous [2] Molecular Architecture of the rotary motor in ATP synthase [3] The molecular mechanism of ATP [4] Essentials for ATP synthesis by F1F0 ATP synthases [5] ATP synthesis driven by proton tranport in |