Sandbox Reserved 481
From Proteopedia
| Line 11: | Line 11: | ||
<Structure load='2q5t' size='305' frame='true' align='center' caption='Protein Structure of Cholix Toxin' scene='Insert optional scene name here' /> | <Structure load='2q5t' size='305' frame='true' align='center' caption='Protein Structure of Cholix Toxin' scene='Insert optional scene name here' /> | ||
==='''X-Ray Crystallography'''=== | ==='''X-Ray Crystallography'''=== | ||
| - | To observe the structure, the Cholix Toxin was crystallized by vapor diffusion against reservoirs containing 23% polyethlene gylcol-10,000, 7.5% ethylene glycol, and 0.1 m HERPES. In addition, the reservoirs were kept at a of pH 7.5 and at a temperature of 19°C. About 40 µL of reservior solution containing 1.25 mM NAD+ solution was added to a 2 µL crystal containing group. The NAD+ was allowed to soak into the crystals for approximately 2-3 minutes. The crystals were then transferred to paratone-N for cryoprotection. Then to visualize it, a diffractometer was used.<ref>Jorgensen, R., Purdy, A., Fieldhouse, R., Kimber, M., Bartlett, D., & Merrill, A. (2009). Cholix toxin, a novel adp-ribosylating factor from vibrio cholerae. Journal of Biological Chemistry, 283(16), 10671-10678. </ref> | + | "To observe the structure, the Cholix Toxin was crystallized by vapor diffusion against reservoirs containing 23% polyethlene gylcol-10,000, 7.5% ethylene glycol, and 0.1 m HERPES." <ref>Jorgensen, R. (Artist). (2008). PDB File Cholix toxin, a novel adp-ribosylating factor from vibrio cholerae.. [Web Graphic]. Retrieved from http://www.rcsb.org/pdb/explore/explore.do?structureId=2q5t</ref> In addition, the reservoirs were kept at a of pH 7.5 and at a temperature of 19°C. About 40 µL of reservior solution containing 1.25 mM NAD+ solution was added to a 2 µL crystal containing group. The NAD+ was allowed to soak into the crystals for approximately 2-3 minutes. The crystals were then transferred to paratone-N for cryoprotection. Then to visualize it, a diffractometer was used.<ref>Jorgensen, R., Purdy, A., Fieldhouse, R., Kimber, M., Bartlett, D., & Merrill, A. (2009). Cholix toxin, a novel adp-ribosylating factor from vibrio cholerae. Journal of Biological Chemistry, 283(16), 10671-10678. </ref> |
==='''Protein Structure'''=== | ==='''Protein Structure'''=== | ||
Revision as of 03:45, 3 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
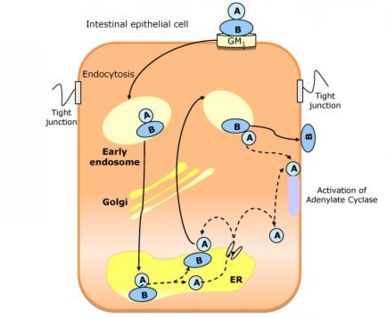
Cholix Toxin from Vibrio choleraeIntroductionCholix toxin (CT) is a class of protein exotoxin originating from the bacteria Vibrio cholerae. It is the third member of eEF2 specific ADP-ribosyltransferase toxins.[1] The toxin uses ADP-riboyltransferase to inactivate eukaryotic elongation factor 2 by transferring ADP-ribose from NAD+ which inhibits protein synthesis and causes cell death. This protein toxin has been known to cause disease in both plants and animals. Cholix is the causative agent of the disease Cholera. It enters eukaryotic cells through endocytosis. Once inside, the toxin transfers an ADP-ribose group to an Arg residue of the GTP binding protein G. This activates adenylate cyclase which leads to an increase amount of cAMP, causing a secretion of Cl-,HCO3-, and water from epithelial cells from the site of colonization [2]. The result is dehydration and loss of electrolytes in infected hosts. "Structurally, it is a three-domain A/B toxin. It consists of a receptor binding domain for receptor mediated endocytosis, a membrane translocation domain for the translocation to the host cytoplasm, and a catalytic domain" [3] Structure
X-Ray Crystallography"To observe the structure, the Cholix Toxin was crystallized by vapor diffusion against reservoirs containing 23% polyethlene gylcol-10,000, 7.5% ethylene glycol, and 0.1 m HERPES." [4] In addition, the reservoirs were kept at a of pH 7.5 and at a temperature of 19°C. About 40 µL of reservior solution containing 1.25 mM NAD+ solution was added to a 2 µL crystal containing group. The NAD+ was allowed to soak into the crystals for approximately 2-3 minutes. The crystals were then transferred to paratone-N for cryoprotection. Then to visualize it, a diffractometer was used.[5] Protein StructureThe sequence of CT is 666 amino acids in length. It contains a total of four disulfide bonds which are located between Cys-43:Cys-47, Cys-240:Cys: 257,Cys-310:Cys-332, and Cys-426:Cys-433.[6]. The secondary structure of the Cholix Toxin consists of 11 and 4 The Cholix Toxin contains three different domains. Though it is not shown, the active site for this toxin is located at Glu-613. interactions with NAD+ include V360,T469,I470,V477,L492, and V500. There is also interactions between the stacking of the nicotinamide ring and Y493 and Y504.[7] Molecule"CT is a bipartite molecule belonging to the AB5 family, which also includes Shiga and pertussis toxins. CT combines one A active subunit and five identical peptides that is assembled into a highly stable pentameric ring named B subunit. The A subunit presents two domains:A1 and A2."[8] The B subunit serves to bind to GM1 ganglioside on the surfaces of intestinal epithelial cells.[9] Both of these subunits are highly essential for the functionality of this toxin. Mechanism of ActionBasic MechanismThe mechanism for the cholix toxin occurs as follows:
ADP-ribosylationNot only does Cholix use ADP-ribosylation to activate the Gs-α subunit of G-Proteins, but it also covalently transfers the ADP-ribose from NAD+ to diphthamide on eEF2 (Eukaryotic Elongation Factor 2) through nucleophilic substitution. This causes for the inhibition of protein synthesis and cell death.[13] Applications
References
|