Sandbox Reserved 455
From Proteopedia
| Line 8: | Line 8: | ||
==Introduction== | ==Introduction== | ||
| - | <Structure load='1te6' size='500' frame='true' align='right' caption='RSCB-Protein Data Bank: Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom' scene=' | + | <Structure load='1te6' size='500' frame='true' align='right' caption='RSCB-Protein Data Bank: Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom' scene='Insert Optional Scene Name Here' /> |
Neuron Specific Enolase (NSE) is one of the five isozymes (isoenzyme) of the [[glycolysis]] enzyme [[enolase]]. An enolase falls under the category of [[lyase]], which is an enzyme that catalyzes the breaking of various chemical bonds by means other than hydrolysis and oxidation, often forming a new double bond or a new ring structure.<ref name="enolase">http://en.wikipedia.org/wiki/Enolase</ref> It is also known as a metalloenzyme, which means it contains a metal ion bound to the protein with one labile coordination site.<ref>http://en.wikipedia.org/wiki/Metalloenzyme#Metalloenzymes</ref> Enolase is present in all tissues and organisms capable of glycolysis or fermentation. It is the ninth and penultimate step of glycolysis, converting 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP). <ref name="enolase" />[[Image:Lobster Enolase .jpg|left|thumb|'''Biological Assembly Image for 1PDY''' Lobster Enolase <ref>X-RAY STRUCTURE AND CATALYTIC MECHANISM OF LOBSTER ENOLASE: http://www.rcsb.org/pdb/explore/explore.do?structureId=1PDY</ref>]] | Neuron Specific Enolase (NSE) is one of the five isozymes (isoenzyme) of the [[glycolysis]] enzyme [[enolase]]. An enolase falls under the category of [[lyase]], which is an enzyme that catalyzes the breaking of various chemical bonds by means other than hydrolysis and oxidation, often forming a new double bond or a new ring structure.<ref name="enolase">http://en.wikipedia.org/wiki/Enolase</ref> It is also known as a metalloenzyme, which means it contains a metal ion bound to the protein with one labile coordination site.<ref>http://en.wikipedia.org/wiki/Metalloenzyme#Metalloenzymes</ref> Enolase is present in all tissues and organisms capable of glycolysis or fermentation. It is the ninth and penultimate step of glycolysis, converting 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP). <ref name="enolase" />[[Image:Lobster Enolase .jpg|left|thumb|'''Biological Assembly Image for 1PDY''' Lobster Enolase <ref>X-RAY STRUCTURE AND CATALYTIC MECHANISM OF LOBSTER ENOLASE: http://www.rcsb.org/pdb/explore/explore.do?structureId=1PDY</ref>]] | ||
| Line 18: | Line 18: | ||
==Structure== | ==Structure== | ||
| - | NSE, as mentioned in the introduction, is composed of the two gamma, γ, subunits of enolase. NSE functions in neurons, but it still performs the duties of glycolytic enolase. Each subunit is composed of two domains, a smaller N-terminal Domain and a larger C-terminal domain. | + | NSE, as mentioned in the introduction, is composed of the two gamma, γ, subunits of enolase. NSE functions in neurons, but it still performs the duties of glycolytic enolase. Each subunit is composed of two domains, a smaller N-terminal Domain and a larger C-terminal domain. For enolase in general, the smaller N-terminal domain consists of three α-helices and four β-sheets. The larger C-terminal domain starts with two β-sheets followed by two α-helices and ends with a barrel composed of alternating β-sheets and α-helices arranged so that the β-beta sheets are surrounded by the α-helices. The enzyme’s compact, globular structure results from significant hydrophobic interactions between these two domains. <ref name="enolase" /> |
| - | + | <scene name='Sandbox_Reserved_455/Active_site_nse/1'>Active Site</scene> | |
| - | + | ||
| - | <scene name='Sandbox_Reserved_455/ | + | |
[[Image:Neuron_Specific_Enolase.jpg|left|thumb|'''Biological Assembly Image for 1TE6''' Enolase 2 <ref>Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom: http://www.rcsb.org/pdb/explore/explore.do?structureId=1te6</ref>]] | [[Image:Neuron_Specific_Enolase.jpg|left|thumb|'''Biological Assembly Image for 1TE6''' Enolase 2 <ref>Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom: http://www.rcsb.org/pdb/explore/explore.do?structureId=1te6</ref>]] | ||
Revision as of 17:56, 8 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Neuron Specific Enolase (Enolase 2)Introduction
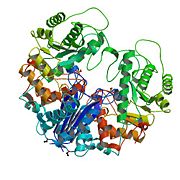 Biological Assembly Image for 1PDY Lobster Enolase [3] Enolase has three subunits (α, β, and γ) and all of these subunits are usually found in vertebrates. Enolase α is ubiquitous, found in all cells, enolase β is muscle-specific, and the γ isozyme is found only in neurons.[4] The subunits can combine in pairs (αα, αβ, αγ, ββ, and γγ) and form the five different isozymes of enolase. It is more common to find the homodimers (αα, ββ, and γγ) in adult human cells. The αα is called enolase 1, the ββ enolase 3, and the γγ enolase 2. Each homodimer still holds their original function: Enolase 1 is non-neuronal enolase (NNE), which is found in a variety of tissues, including liver, brain, kidney, spleen, adipose, enolase 3 is muscle specific enolase (MSE), and enolase 2 neuron-specific enolase (NSE). [1] NSE (also called Gamma-Enolase) is the most abundant form of the glycolytic enolase found in adult neurons and is thought to serve as a growth factor in neurons. NSE is useful in studying neuronal differentiation and is, therefore, a valuable tool for visualizing the entire neuron and endocrine systems. [5] NSE is mainly found in mammals, and it functions as a Phosphopyruvate dehydratase. It is specifically a hydro-lyase, which cleave carbon-oxygen bonds. In humans, NSE is encoded by the ENO2 gene.[6] The systematic name of this enzyme class is 2-phospho-D-glycerate hydro-lyase (phosphoenolpyruvate-forming). Other names in common use include nervous-system specific enolase, phosphoenolpyruvate hydrates, 2-phosphoglycerate dehydrates, 2-phosphoglyceric dehydrates, gamma-enolase, and 2-phospho-D-glycerate hydro-lyase. The only inhibitor known for the enzyme so far is Phosphonoacetohydroxamate [7] StructureNSE, as mentioned in the introduction, is composed of the two gamma, γ, subunits of enolase. NSE functions in neurons, but it still performs the duties of glycolytic enolase. Each subunit is composed of two domains, a smaller N-terminal Domain and a larger C-terminal domain. For enolase in general, the smaller N-terminal domain consists of three α-helices and four β-sheets. The larger C-terminal domain starts with two β-sheets followed by two α-helices and ends with a barrel composed of alternating β-sheets and α-helices arranged so that the β-beta sheets are surrounded by the α-helices. The enzyme’s compact, globular structure results from significant hydrophobic interactions between these two domains. [1]
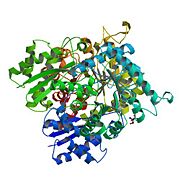 Biological Assembly Image for 1TE6 Enolase 2 [8] Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps MechanismLet's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps MedicineLet's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps Let's see how the text wraps References
|
