Alcohol dehydrogenase from Entamoeba histolytica
From Proteopedia
(New page: 200px <!-- The line below this paragraph, containing "STRUCTURE_1y9a", creates the "Structure Box" on the page. You may change the PDB parameter (which sets the PD...) |
|||
| Line 9: | Line 9: | ||
{{STRUCTURE_1y9a| PDB=1y9a | SCENE= }} | {{STRUCTURE_1y9a| PDB=1y9a | SCENE= }} | ||
| - | ===Alcohol Dehydrogenase from Entamoeba histolotica | + | ===Alcohol Dehydrogenase from ''Entamoeba histolotica'' (EhADH1)=== |
(see also [[Tetrameric alcohol dehydrogenases]]) | (see also [[Tetrameric alcohol dehydrogenases]]) | ||
| Line 21: | Line 21: | ||
<applet load='2oui' size='500' frame='true' align='right' scene='2oui/Common_view/2' /> | <applet load='2oui' size='500' frame='true' align='right' scene='2oui/Common_view/2' /> | ||
| - | <scene name='2oui/Tet/5'>Superposition</scene> of the structures of the <font color='lime'><b>wild-type apo-EhADH1 (colored lime</b></font>, [[1y9a]]) and the <font color='orange'><b>apo D275P-EhADH1 mutant (colored orange)</b></font> ([[2oui]]). <font color='red'><b>Pro275 and Asp275 are labeled red.</b></font> Residues within a distance of 4 Å from the mutation are shown (names of monomers are in brackets). Replacing <scene name='2oui/Tet/8'>Asp275</scene> with <scene name='2oui/Tet/7'>Pro</scene> significantly enhanced the thermal stability of EhADH1: ΔT<sub>1/2</sub><sup>60min</sup> = +9.3°C, ΔT<sub>1/2</sub><sup>CD</sup> = +10°C. The reverse mutation in the thermophilic <scene name='Tetrameric_alcohol_dehydrogenases/Mut/3'>TbADH</scene> ([[1ykf]]; <font color='magenta'><b>colored magenta</b></font>) - substitution of wt TbADH Pro275 with <scene name='Tetrameric_alcohol_dehydrogenases/Mut/2'>Asp</scene> ([[2nvb]]; <font color='cyan'><b>colored cyan</b></font>) reduced the thermal stability of the enzyme: ΔT<sub>1/2</sub><sup>60min</sup> = -13.8°C, ΔT<sub>1/2</sub><sup>CD</sup> = -18.8°C. Nitrogen and oxygen atoms are colored in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. <font color='red'><b>Pro275 and Asp275 are labeled red</b></font> (names of monomers are in brackets). These findings indicate that a single proline mutation is responsible for the significant differences in the thermal stability of ADHs, and show the importance of prolines in the protein stability. It was also shown that substitution by proline at the important positions could significantly stabilize the protein. | + | <scene name='2oui/Tet/5'>Superposition</scene> of the structures of the <font color='lime'><b>wild-type apo-EhADH1 (alcohol dehydrogenase from ''Entamoeba histolotica'', colored lime</b></font>, [[1y9a]]) and the <font color='orange'><b>apo D275P-EhADH1 mutant (colored orange)</b></font> ([[2oui]]). <font color='red'><b>Pro275 and Asp275 are labeled red.</b></font> Residues within a distance of 4 Å from the mutation are shown (names of monomers are in brackets). Replacing <scene name='2oui/Tet/8'>Asp275</scene> with <scene name='2oui/Tet/7'>Pro</scene> significantly enhanced the thermal stability of EhADH1: ΔT<sub>1/2</sub><sup>60min</sup> = +9.3°C, ΔT<sub>1/2</sub><sup>CD</sup> = +10°C. The reverse mutation in the thermophilic alcohol dehydrogenase from ''Thermoanaerobacter brockii'' <scene name='Tetrameric_alcohol_dehydrogenases/Mut/3'>TbADH</scene> ([[1ykf]]; <font color='magenta'><b>colored magenta</b></font>) - substitution of wt TbADH Pro275 with <scene name='Tetrameric_alcohol_dehydrogenases/Mut/2'>Asp</scene> ([[2nvb]]; <font color='cyan'><b>colored cyan</b></font>) reduced the thermal stability of the enzyme: ΔT<sub>1/2</sub><sup>60min</sup> = -13.8°C, ΔT<sub>1/2</sub><sup>CD</sup> = -18.8°C. Nitrogen and oxygen atoms are colored in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. <font color='red'><b>Pro275 and Asp275 are labeled red</b></font> (names of monomers are in brackets). These findings indicate that a single proline mutation is responsible for the significant differences in the thermal stability of ADHs, and show the importance of prolines in the protein stability. It was also shown that substitution by proline at the important positions could significantly stabilize the protein. |
==About this Structure== | ==About this Structure== | ||
1Y9A is a 2 chains structure of sequences from [http://en.wikipedia.org/wiki/Entamoeba_histolytica Entamoeba histolytica]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Y9A OCA]. | 1Y9A is a 2 chains structure of sequences from [http://en.wikipedia.org/wiki/Entamoeba_histolytica Entamoeba histolytica]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Y9A OCA]. | ||
Revision as of 09:50, 6 June 2012
Alcohol Dehydrogenase from Entamoeba histolotica (EhADH1)
(see also Tetrameric alcohol dehydrogenases)
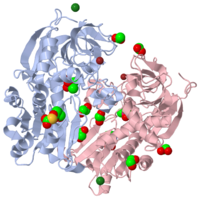
The structure of the apo form of alcohol dehydrogenase from a single-cell eukaryotic source, Entamoeba histolytica, has been determined at 1.8 A. To date, bacterial and archeal alcohol dehydrogenases, which are biologically active as tetramers, have crystallized with tetramers in the asymmetric unit. However, the current structure has one independent dimer per asymmetric unit and the full tetramer is generated by application of the crystallographic twofold symmetry element. This structure reveals that many of the crystallization and cryoprotection components, such as cacodylate, ethylene glycol, zinc ions and acetate, have been incorporated. These crystallization solution elements are found within the molecule and at the packing interfaces as an integral part of the three-dimensional arrangements of the tetramers. In addition, an unexpected modification of aspartic acid to O-carboxysulfanyl-4-oxo-L-homoserine was found at residue 245.
Structure of alcohol dehydrogenase from Entamoeba histolytica., Shimon LJ, Goihberg E, Peretz M, Burstein Y, Frolow F, Acta Crystallogr D Biol Crystallogr. 2006 May;62(Pt 5):541-7. Epub 2006, Apr 19. PMID:16627948
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
|
of the structures of the wild-type apo-EhADH1 (alcohol dehydrogenase from Entamoeba histolotica, colored lime, 1y9a) and the apo D275P-EhADH1 mutant (colored orange) (2oui). Pro275 and Asp275 are labeled red. Residues within a distance of 4 Å from the mutation are shown (names of monomers are in brackets). Replacing with significantly enhanced the thermal stability of EhADH1: ΔT1/260min = +9.3°C, ΔT1/2CD = +10°C. The reverse mutation in the thermophilic alcohol dehydrogenase from Thermoanaerobacter brockii (1ykf; colored magenta) - substitution of wt TbADH Pro275 with (2nvb; colored cyan) reduced the thermal stability of the enzyme: ΔT1/260min = -13.8°C, ΔT1/2CD = -18.8°C. Nitrogen and oxygen atoms are colored in CPK colors. Pro275 and Asp275 are labeled red (names of monomers are in brackets). These findings indicate that a single proline mutation is responsible for the significant differences in the thermal stability of ADHs, and show the importance of prolines in the protein stability. It was also shown that substitution by proline at the important positions could significantly stabilize the protein.
About this Structure
1Y9A is a 2 chains structure of sequences from Entamoeba histolytica. Full crystallographic information is available from OCA.
Reference
- Shimon LJ, Goihberg E, Peretz M, Burstein Y, Frolow F. Structure of alcohol dehydrogenase from Entamoeba histolytica. Acta Crystallogr D Biol Crystallogr. 2006 May;62(Pt 5):541-7. Epub 2006, Apr 19. PMID:16627948 doi:10.1107/S0907444906009292