This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Transcription-repair coupling factor
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='2eyq' size='350' side='right' scene='2eyq/Asymmunit/2' caption=''> | ||
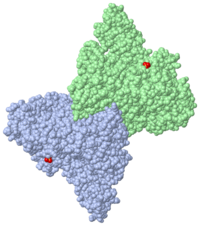
[[Image:2eyq.png|left|200px|thumb|Crystal Structure of Transcription-repair coupling factor [[2eyq]]]] | [[Image:2eyq.png|left|200px|thumb|Crystal Structure of Transcription-repair coupling factor [[2eyq]]]] | ||
| - | <StructureSection load='2eyq' size='350' side='right' scene='2eyq/Asymmunit/2' caption='2eyq asymmetric unit, resolution 3.20Å. <br> Click <scene name='2eyq/Biolunit/1'>here</scene> to display biological molecule'> | ||
'''Transcription-repair coupling factor (TRCF)''' enables the coupling of these processes in bacteria and humans. The TRCF has ATPase activity. <scene name='2eyq/Asymmunit/2'>The crystallographic asymmetric unit of the solved structure contains 2 monomers of Mfd (residues 2-1147 of 1151 for one molecule and 5-1147 of the other), five HEPES molecules, and the sulfate ions.</scene> | '''Transcription-repair coupling factor (TRCF)''' enables the coupling of these processes in bacteria and humans. The TRCF has ATPase activity. <scene name='2eyq/Asymmunit/2'>The crystallographic asymmetric unit of the solved structure contains 2 monomers of Mfd (residues 2-1147 of 1151 for one molecule and 5-1147 of the other), five HEPES molecules, and the sulfate ions.</scene> | ||
Revision as of 10:34, 7 June 2012
| |||||||||||
Reference
- Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006 Feb 10;124(3):507-20. PMID:16469698 doi:10.1016/j.cell.2005.11.045
Created with the participation of Wayne Decatur
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Alexander Berchansky, Wayne Decatur