Allosteric modulation of H-Ras GTPase
From Proteopedia
(→Mutations) |
|||
| Line 6: | Line 6: | ||
Mutation is Q61 causes shift in amino acids | Mutation is Q61 causes shift in amino acids | ||
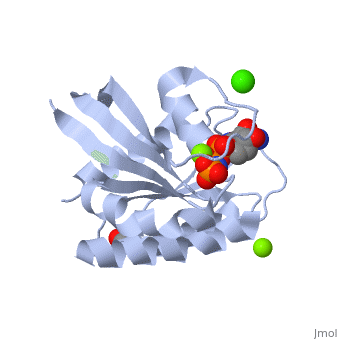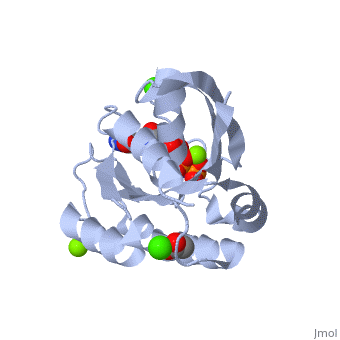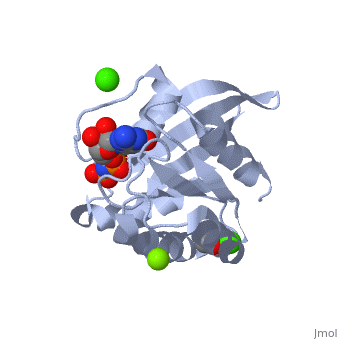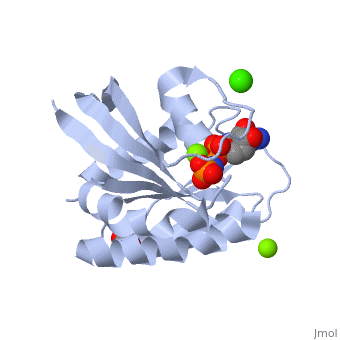
| - | [[Image:RasCacliumacetate.jpg| | + | [[Image:RasCacliumacetate.jpg|400px|center|Calcium Acetate/Loop 7/Helix 3/GppNHp]] |
Revision as of 02:15, 19 June 2012
| |||||||||
| 3k8y, resolution 1.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Gene: | HRAS, HRAS1 (Homo sapiens) | ||||||||
| Related: | 2rge, 3k9n, 3k9l | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
H-Ras is a small monomeric GTPase protein that is found inside of cells. It helps regulate cell division in cells through signal transduction pathways such as the Ras pathway. H-Ras is also known as a molecular switch, it contains two switch regions: Switch I and Switch II. Switch I is a Thymine 35 and switch II is a Glycine 60, both of these switches depend on the phosphate group on a GTP molecule to turn “on” and “off”. In the Ras catalytic cycle, Ras is activated when it attaches to a GTP molecule turning both switch regions “on”. This pathway also involves GTPase activating protein, which hydrolyzes the GTP attached to the Ras causing the switches to turn “off”.
Mutations
Mutation is Q61 causes shift in amino acids