Alcohol dehydrogenase from Entamoeba histolytica
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
[[Image:1y9a.png|left|200px]] | [[Image:1y9a.png|left|200px]] | ||
| - | ===Alcohol Dehydrogenase from ''Entamoeba | + | ===Alcohol Dehydrogenase from ''Entamoeba histolytica'' (EhADH1)=== |
(see also [[Tetrameric alcohol dehydrogenases]]) | (see also [[Tetrameric alcohol dehydrogenases]]) | ||
| Line 9: | Line 9: | ||
{{Clear}} | {{Clear}} | ||
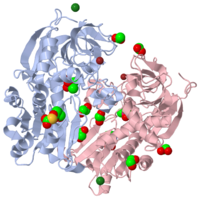
| - | <scene name='2oui/Tet/5'>Superposition</scene> of the structures of the <font color='lime'><b>wild-type apo-EhADH1 (alcohol dehydrogenase from ''Entamoeba | + | <scene name='2oui/Tet/5'>Superposition</scene> of the structures of the <font color='lime'><b>wild-type apo-EhADH1 (alcohol dehydrogenase from ''Entamoeba histolytica'', colored lime</b></font>, [[1y9a]]) and the <font color='orange'><b>apo D275P-EhADH1 mutant (colored orange)</b></font> ([[2oui]]). <font color='red'><b>Pro275 and Asp275 are labeled red.</b></font> Residues within a distance of 4 Å from the mutation are shown (names of monomers are in brackets). Replacing <scene name='2oui/Tet/8'>Asp275</scene> with <scene name='2oui/Tet/7'>Pro</scene> significantly enhanced the thermal stability of EhADH1: ΔT<sub>1/2</sub><sup>60min</sup> = +9.3°C, ΔT<sub>1/2</sub><sup>CD</sup> = +10°C. The reverse mutation in the thermophilic alcohol dehydrogenase from ''Thermoanaerobacter brockii'' <scene name='Tetrameric_alcohol_dehydrogenases/Mut/3'>TbADH</scene> ([[1ykf]]; <font color='magenta'><b>colored magenta</b></font>) - substitution of wt TbADH Pro275 with <scene name='Tetrameric_alcohol_dehydrogenases/Mut/2'>Asp</scene> ([[2nvb]]; <font color='cyan'><b>colored cyan</b></font>) reduced the thermal stability of the enzyme: ΔT<sub>1/2</sub><sup>60min</sup> = -13.8°C, ΔT<sub>1/2</sub><sup>CD</sup> = -18.8°C. Nitrogen and oxygen atoms are colored in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. <font color='red'><b>Pro275 and Asp275 are labeled red</b></font> (names of monomers are in brackets). These findings indicate that a single proline mutation is responsible for the significant differences in the thermal stability of ADHs, and show the importance of prolines in the protein stability. It was also shown that substitution by proline at the important positions could significantly stabilize the protein. |
==About this Structure== | ==About this Structure== | ||
1Y9A is a 2 chains structure of sequences from [http://en.wikipedia.org/wiki/Entamoeba_histolytica Entamoeba histolytica]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Y9A OCA]. | 1Y9A is a 2 chains structure of sequences from [http://en.wikipedia.org/wiki/Entamoeba_histolytica Entamoeba histolytica]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Y9A OCA]. | ||
Revision as of 08:06, 15 July 2012
| |||||||||||
Reference
- Shimon LJ, Goihberg E, Peretz M, Burstein Y, Frolow F. Structure of alcohol dehydrogenase from Entamoeba histolytica. Acta Crystallogr D Biol Crystallogr. 2006 May;62(Pt 5):541-7. Epub 2006, Apr 19. PMID:16627948 doi:10.1107/S0907444906009292