Prp8
From Proteopedia
| Line 26: | Line 26: | ||
The proline rich region is present at the N-terminus of Prp8, approximately running from residues 5 - 78 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. This proline tract is found at the N-terminus of all fungal Prp8 sequences, and two rice sequences but is absent from most other organisms <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The proline rich region usually adopts an extended helical structure with three residues per turn <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The [http://en.wikipedia.org/wiki/Nuclear_localization_sequence nuclear localization signal] is also at the N-terminus of the protein, extending through amino acid residues 81 - 120, and can be located in the first 500 amino acids of the majority of organisms' Prp8 sequences <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The nuclear localization signal contains two clusters of positively charged amino acid residues which are separated by a variable region of 10 - 12 amino acids, and organisms lacking this classical signal motif usually possess an alternative within the N-terminus of their Prp8 <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The bromodomain is the last major region that is present at the N-terminus of Prp8, running from residues 200 - 315 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. It possesses low sequence identity (<14%) with commonplace bromodomains, but appears to contain no insertions or deletions despite its low identity with other sequences <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. The domain consists of four alpha-helices and two loops which generally interact with and recognize lysine residues <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. The recognition is facilitated by several highly conserved amino acids that stabilize the helix bundle <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. | The proline rich region is present at the N-terminus of Prp8, approximately running from residues 5 - 78 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. This proline tract is found at the N-terminus of all fungal Prp8 sequences, and two rice sequences but is absent from most other organisms <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The proline rich region usually adopts an extended helical structure with three residues per turn <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The [http://en.wikipedia.org/wiki/Nuclear_localization_sequence nuclear localization signal] is also at the N-terminus of the protein, extending through amino acid residues 81 - 120, and can be located in the first 500 amino acids of the majority of organisms' Prp8 sequences <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The nuclear localization signal contains two clusters of positively charged amino acid residues which are separated by a variable region of 10 - 12 amino acids, and organisms lacking this classical signal motif usually possess an alternative within the N-terminus of their Prp8 <ref name='Prp8: At the heart of the spliceosome'>DOI:10.1261/rna.2220705</ref>. The bromodomain is the last major region that is present at the N-terminus of Prp8, running from residues 200 - 315 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. It possesses low sequence identity (<14%) with commonplace bromodomains, but appears to contain no insertions or deletions despite its low identity with other sequences <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. The domain consists of four alpha-helices and two loops which generally interact with and recognize lysine residues <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. The recognition is facilitated by several highly conserved amino acids that stabilize the helix bundle <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. | ||
| - | The reverse transcriptase-like domain is located approximately halfway through Prp8 and extends through residues 950 - 1220 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. This domain is only reverse transcriptase-like because it does contain the conventional core set of alpha-helices and beta-strands that are a general characteristic of RT domains, but lacks key motifs that would normally confer its catalytic activity <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. However, the Prp8 RT-like domain still possesses the potential to bind RNA <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. Prp8 also contains an RNase H-like domain towards its C-terminal end (residues 1836 - 2092) <ref name='Mechanism for aar2p function as a U5 snRNP assembly factor'>PMID:21764848</ref>. The central region of the RNase H-like domain consists of a mixed alpha/beta fold, which resembles the overall shape of a mitten <ref name='Mechanism for aar2p function as a U5 snRNP assembly factor'>PMID:21764848</ref>. Following this analogy, the central 6 stranded beta sheets and alpha helices are the palm with a beta hairpin as the thumb and alpha helices as fingers. This is represented by the portion of the PDB figure that | + | The reverse transcriptase-like domain is located approximately halfway through Prp8 and extends through residues 950 - 1220 <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. This domain is only reverse transcriptase-like because it does contain the conventional core set of alpha-helices and beta-strands that are a general characteristic of RT domains, but lacks key motifs that would normally confer its catalytic activity <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. However, the Prp8 RT-like domain still possesses the potential to bind RNA <ref name='Prp8, the pivotal protein of the spliceosomal catalytic center evolved from a retroelement-encoded reverse transcriptase'>DOI:10.1261/rna.2396011</ref>. Prp8 also contains an RNase H-like domain towards its C-terminal end (residues 1836 - 2092) <ref name='Mechanism for aar2p function as a U5 snRNP assembly factor'>PMID:21764848</ref>. The central region of the RNase H-like domain consists of a mixed alpha/beta fold, which resembles the overall shape of a mitten <ref name='Mechanism for aar2p function as a U5 snRNP assembly factor'>PMID:21764848</ref>. Following this analogy, the central 6 stranded beta sheets and alpha helices are the palm with a beta hairpin as the thumb and alpha helices as fingers. This is represented by the portion of the PDB figure that has yellow apha helices with blue beta sheets and the blue unstructured protein chain. The groove created by the mitten like structure is lined with positive surface potential but also lacks key residues for RNase activity which is perfect for binding RNA, and has a very similar structure to the reverse transcriptase-like domain mentioned above <ref name='Structure and function of an RNase H domain at the heart of the spliceosome'>DOI:10.1038/emboj.2008.209</ref>. The RNase domain possesses conserved charged residues at D1853/1781, D1854/1782, and Q1907/1835 (yeast/human numbering) which are thought to mediate interactions with the negatively charged backbone of an RNA molecule <ref name='Structure and function of an RNase H domain at the heart of the spliceosome'>DOI:10.1038/emboj.2008.209</ref>. Finally, the final domain one the C-terminal end of Prp8 is a metalloprotease/Jab1-like domain which extends through residues 2112 - 2413 and is represented by |
=Function of Prp8= | =Function of Prp8= | ||
Revision as of 05:00, 23 July 2012
Contents |
Introduction
Prp8 Background
Prp8's name originates from the discovery that it is intricately involved with pre-m RNA processing within Eukaryotic organisms, and extensive research on the protein has revealed it is indispensable for the catalytic activity of the spliceosome earning it the nickname of 'Master Regulator of the Spliceosome' [1]. UV crosslinking experiments have shown that Prp8 has contact with the 5' splice site, 3' splice site, polypyrimidine tract, and branch point of pre-mRNA transcripts, as well as associating with all five of the snRNAs and their associated proteins [2]. Thus, Prp8 is the only protein to date involved with pre-mRNA splicing that actually contacts all catalytic elements of the spliceosome, indicating that it indeed has a very crucial role in pre-mRNA splicing catalysis, and resides in the catalytic heart of the splicing comlex. Two other unique properties of Prp8 are its size and evolutionary conservation. Prp8 is the largest of all the spliceosomal proteins at approximately 250 kDa and is 2317 - 2416 amino acid residues in length (depending on the orthologue) [1]. In all organisms that exhibit Prp8, it is always found as a U5 snRNP component and is highly conserved across species, with >60% identity between fungi, mammals, and plants [3]. Although Prp8 is highly conserved between species, its origin is shrouded in mystery because it contains no sequence homology to one family of proteins, but instead exhibits structural properties and domains from a range of conserved protein families while adding a twist to each that is Prp8 specific [1]. Even though Prp8 has been under extensive investigation for over a decade, its specific molecular mechanisms and functions are still unknown, but in light of all the information available, a hypothesis on Prp8's function can be postulated; Prp8 definitely acts as a large scaffold to coordinate the spliceosome's activity because the protein interacts with every part of the complex, however it is unclear whether Prp8 plays a direct role in splicing catalysis or only acts as a guide allowing for correct orientation and dynamic movements [1].
Pre-mRNA Splicing
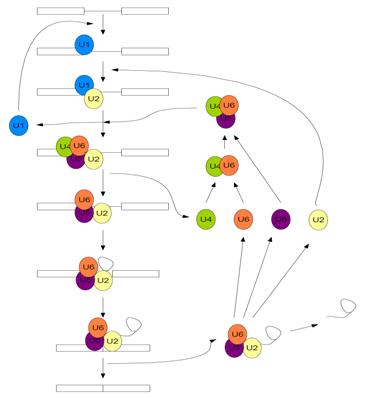
Pre-mRNA splicing is carried out by one of the most complex pieces of cellular machinery known to date; the Spliceosome. It is a large, catalytic protein-RNA complex responsible for removing the intronic sequences of pre-mRNA and “splicing” together the exonic sequences to form mature mRNA. The spliceosome is made up of approximately 145 distinct spliceosomal proteins, and five small nuclear RNAs (snRNAs) which each have a distinct set of proteins that interact with them forming snRNPs [5]. The five snRNAs have been subsequently named U1, U2, U4, U5, and U6. The spliceosome carries out a two-step transesterification reaction through a series of chemical steps in order to remove the intronic sequences from the pre-mRNA, which is facilitated by the movements, rearrangements, and dynamic exchanges of the snRNAs and associated splicing proteins [4].
First, the U1 snRNP recognizes and binds to the 5’ splice site of the pre-mRNA and the U2 snRNP recognizes and binds to the branch site of the intronic sequence, which contains a conserved adenine nucleotide, which is collectively known as complex A [6]. Subsequently, the U4/U6 U5 tri-snRNP moves in and associates with the intronic sequence (now known as complex B)[6]. This causes the U4 and U1 snRNPs to be displaced, forming the catalytically active spliceosome (complex B*) and the remaining U2, U5, and U6 snRNP facilitate the two transesterification reactions, dissociate from the now mature mRNA, and are recycled for subsequent use in another splicing reaction [6].
As stated above, the chemical mechanism pre-mRNA splicing follows is a two-step transesterification reaction. With the release of U1 and U4, following the subsequent rearrangement of the remaining snRNPs, the 2’ hydroxyl of the conserved adenine nucleotide carries out a nucleophilic attack on the 5’ splice site, which cuts the backbone of the mRNA, freeing the first exon [4]. Now the 5’ end of the intron is covalently bonded to the adenine nucleotide, forming a lariat structure [4]. Finally, the 3’ OH of the first exon nucleophilically attacks the 5’ end of the second exon, displacing the intron and splicing the exons together [4]. This is followed by the subsequent release of the lariat structure and the remaining spliceosome dissociates [4].
Structure of Prp8
|
Prp8's structure can be summarized as an amalgamation of domains from a variety of proteins, each of which has been slightly modified from their conventional forms and are now unique to Prp8. The major features present in Prp8 are a proline rich region, nuclear localization signal, bromodomain, reverse transcriptase-like domain, RNase H-like domain, and finally a metalloprotease/Jab1-like domain [7].
The proline rich region is present at the N-terminus of Prp8, approximately running from residues 5 - 78 [7]. This proline tract is found at the N-terminus of all fungal Prp8 sequences, and two rice sequences but is absent from most other organisms [1]. The proline rich region usually adopts an extended helical structure with three residues per turn [1]. The nuclear localization signal is also at the N-terminus of the protein, extending through amino acid residues 81 - 120, and can be located in the first 500 amino acids of the majority of organisms' Prp8 sequences [1]. The nuclear localization signal contains two clusters of positively charged amino acid residues which are separated by a variable region of 10 - 12 amino acids, and organisms lacking this classical signal motif usually possess an alternative within the N-terminus of their Prp8 [1]. The bromodomain is the last major region that is present at the N-terminus of Prp8, running from residues 200 - 315 [7]. It possesses low sequence identity (<14%) with commonplace bromodomains, but appears to contain no insertions or deletions despite its low identity with other sequences [7]. The domain consists of four alpha-helices and two loops which generally interact with and recognize lysine residues [7]. The recognition is facilitated by several highly conserved amino acids that stabilize the helix bundle [7].
The reverse transcriptase-like domain is located approximately halfway through Prp8 and extends through residues 950 - 1220 [7]. This domain is only reverse transcriptase-like because it does contain the conventional core set of alpha-helices and beta-strands that are a general characteristic of RT domains, but lacks key motifs that would normally confer its catalytic activity [7]. However, the Prp8 RT-like domain still possesses the potential to bind RNA [7]. Prp8 also contains an RNase H-like domain towards its C-terminal end (residues 1836 - 2092) [8]. The central region of the RNase H-like domain consists of a mixed alpha/beta fold, which resembles the overall shape of a mitten [8]. Following this analogy, the central 6 stranded beta sheets and alpha helices are the palm with a beta hairpin as the thumb and alpha helices as fingers. This is represented by the portion of the PDB figure that has yellow apha helices with blue beta sheets and the blue unstructured protein chain. The groove created by the mitten like structure is lined with positive surface potential but also lacks key residues for RNase activity which is perfect for binding RNA, and has a very similar structure to the reverse transcriptase-like domain mentioned above [9]. The RNase domain possesses conserved charged residues at D1853/1781, D1854/1782, and Q1907/1835 (yeast/human numbering) which are thought to mediate interactions with the negatively charged backbone of an RNA molecule [9]. Finally, the final domain one the C-terminal end of Prp8 is a metalloprotease/Jab1-like domain which extends through residues 2112 - 2413 and is represented by
Function of Prp8
Evolution of Prp8
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005 May;11(5):533-57. PMID:15840809 doi:10.1261/rna.2220705
- ↑ Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999 Aug 1;13(15):1983-93. PMID:10444596
- ↑ Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011 May;17(5):799-808. Epub 2011 Mar 25. PMID:21441348 doi:10.1261/rna.2396011
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157-63. PMID:1853200
- ↑ Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002 Sep 12;419(6903):182-5. PMID:12226669 doi:10.1038/nature01031
- ↑ 6.0 6.1 6.2 Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559-99. PMID:1839712 doi:http://dx.doi.org/10.1146/annurev.cb.07.110191.003015
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011 May;17(5):799-808. Epub 2011 Mar 25. PMID:21441348 doi:10.1261/rna.2396011
- ↑ 8.0 8.1 Weber G, Cristao VF, Alves FD, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev. 2011 Jul 15. PMID:21764848 doi:10.1101/gad.635911
- ↑ 9.0 9.1 Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J. 2008 Nov 5;27(21):2929-40. Epub 2008 Oct 9. PMID:18843295 doi:10.1038/emboj.2008.209
