1q21
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1q21.png|left|200px]] | [[Image:1q21.png|left|200px]] | ||
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_1q21", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
{{STRUCTURE_1q21| PDB=1q21 | SCENE= }} | {{STRUCTURE_1q21| PDB=1q21 | SCENE= }} | ||
===CRYSTAL STRUCTURES AT 2.2 ANGSTROMS RESOLUTION OF THE CATALYTIC DOMAINS OF NORMAL RAS PROTEIN AND AN ONCOGENIC MUTANT COMPLEXED WITH GSP=== | ===CRYSTAL STRUCTURES AT 2.2 ANGSTROMS RESOLUTION OF THE CATALYTIC DOMAINS OF NORMAL RAS PROTEIN AND AN ONCOGENIC MUTANT COMPLEXED WITH GSP=== | ||
| - | |||
| - | <!-- | ||
| - | The line below this paragraph, {{ABSTRACT_PUBMED_1899707}}, adds the Publication Abstract to the page | ||
| - | (as it appears on PubMed at http://www.pubmed.gov), where 1899707 is the PubMed ID number. | ||
| - | --> | ||
{{ABSTRACT_PUBMED_1899707}} | {{ABSTRACT_PUBMED_1899707}} | ||
==About this Structure== | ==About this Structure== | ||
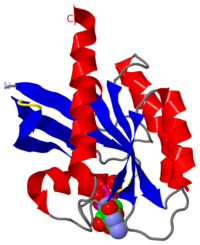
| - | [[1q21]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=2p21 2p21]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Q21 OCA]. | + | [[1q21]] is a 1 chain structure of [[GTPase HRas]] with sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=2p21 2p21]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1Q21 OCA]. |
==See Also== | ==See Also== | ||
| - | *[[ | + | *[[GTPase HRas|GTPase HRas]] |
==Reference== | ==Reference== | ||
| - | <ref group="xtra">PMID: | + | <ref group="xtra">PMID:001899707</ref><ref group="xtra">PMID:009398520</ref><ref group="xtra">PMID:009428770</ref><references group="xtra"/> |
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Kim, S H.]] | [[Category: Kim, S H.]] | ||
[[Category: Oncogene protein]] | [[Category: Oncogene protein]] | ||
Revision as of 12:51, 25 July 2012
| |||||||||
| 1q21, resolution 2.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
CRYSTAL STRUCTURES AT 2.2 ANGSTROMS RESOLUTION OF THE CATALYTIC DOMAINS OF NORMAL RAS PROTEIN AND AN ONCOGENIC MUTANT COMPLEXED WITH GSP
The biological functions of ras proteins are controlled by the bound guanine nucleotide GDP or GTP. The GTP-bound conformation is biologically active, and is rapidly deactivated to the GDP-bound conformation through interaction with GAP (GTPase Activating Protein). Most transforming mutants of ras proteins have drastically reduced GTP hydrolysis rates even in the presence of GAP. The crystal structures of the GDP complexes of ras proteins at 2.2 A resolution reveal the detailed interaction between the ras proteins and the GDP molecule. All the currently known transforming mutation positions are clustered around the bound guanine nucleotide molecule. The presumed "effector" region and the GAP recognition region are both highly exposed. No significant structural differences were found between the GDP complexes of normal ras protein and the oncogenic mutant with valine at position 12, except the side-chain of the valine residue. However, comparison with GTP-analog complexes of ras proteins suggests that the valine side-chain may inhibit GTP hydrolysis in two possible ways: (1) interacting directly with the gamma-phosphate and altering its orientation or the conformation of protein residues around the phosphates; and/or (2) preventing either the departure of gamma-phosphate on GTP hydrolysis or the entrance of a nucleophilic group to attack the gamma-phosphate. The structural similarity between ras protein and the bacterial elongation factor Tu suggests that their common structural motif might be conserved for other guanine nucleotide binding proteins.
Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP., Tong LA, de Vos AM, Milburn MV, Kim SH, J Mol Biol. 1991 Feb 5;217(3):503-16. PMID:1899707
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
1q21 is a 1 chain structure of GTPase HRas with sequence from Homo sapiens. This structure supersedes the now removed PDB entry 2p21. Full crystallographic information is available from OCA.
See Also
Reference
- Tong LA, de Vos AM, Milburn MV, Kim SH. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991 Feb 5;217(3):503-16. PMID:1899707
- Ma J, Karplus M. Ligand-induced conformational changes in ras p21: a normal mode and energy minimization analysis. J Mol Biol. 1997 Nov 21;274(1):114-31. PMID:9398520 doi:10.1006/jmbi.1997.1313
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998 Jan 8;391(6663):203-6. PMID:9428770 doi:10.1038/34472