This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1ecr
From Proteopedia
(New page: 200px<br /><applet load="1ecr" size="450" color="white" frame="true" align="right" spinBox="true" caption="1ecr, resolution 2.700Å" /> '''ESCHERICHIA COLI RE...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1ecr.gif|left|200px]]<br /><applet load="1ecr" size=" | + | [[Image:1ecr.gif|left|200px]]<br /><applet load="1ecr" size="350" color="white" frame="true" align="right" spinBox="true" |
caption="1ecr, resolution 2.700Å" /> | caption="1ecr, resolution 2.700Å" /> | ||
'''ESCHERICHIA COLI REPLICATION TERMINATOR PROTEIN (TUS) COMPLEXED WITH DNA'''<br /> | '''ESCHERICHIA COLI REPLICATION TERMINATOR PROTEIN (TUS) COMPLEXED WITH DNA'''<br /> | ||
==Overview== | ==Overview== | ||
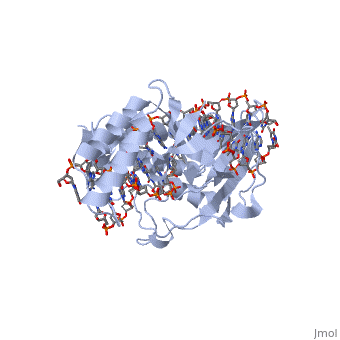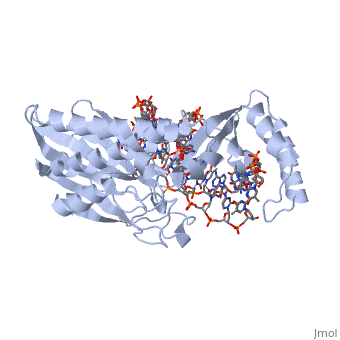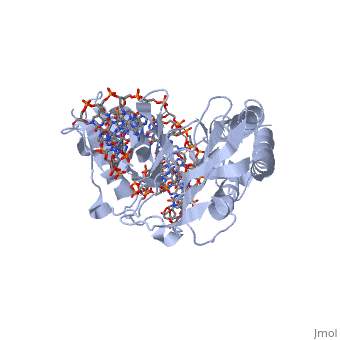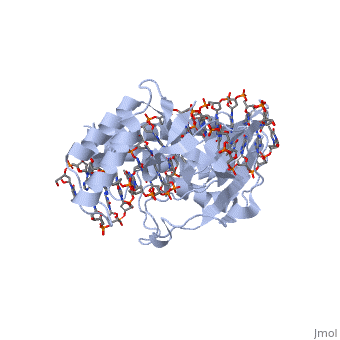
| - | The crystal structure of the Escherichia coli replication-terminator | + | The crystal structure of the Escherichia coli replication-terminator protein (Tus) bound to terminus-site (Ter) DNA has been determined at 2.7 A resolution. The Tus protein folds into a previously undescribed architecture divided into two domains by a central basic cleft. This cleft accommodates locally deformed B-form Ter DNA and makes extensive contacts with the major groove, mainly through two interdomain beta-strands. The unusual structural features of this complex may explain how the replication fork is halted in only one direction. |
==About this Structure== | ==About this Structure== | ||
| - | 1ECR is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http:// | + | 1ECR is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1ECR OCA]. |
==Reference== | ==Reference== | ||
| Line 19: | Line 19: | ||
[[Category: dna-binding]] | [[Category: dna-binding]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 12:26:21 2008'' |
Revision as of 10:26, 21 February 2008
|
ESCHERICHIA COLI REPLICATION TERMINATOR PROTEIN (TUS) COMPLEXED WITH DNA
Overview
The crystal structure of the Escherichia coli replication-terminator protein (Tus) bound to terminus-site (Ter) DNA has been determined at 2.7 A resolution. The Tus protein folds into a previously undescribed architecture divided into two domains by a central basic cleft. This cleft accommodates locally deformed B-form Ter DNA and makes extensive contacts with the major groove, mainly through two interdomain beta-strands. The unusual structural features of this complex may explain how the replication fork is halted in only one direction.
About this Structure
1ECR is a Single protein structure of sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
Structure of a replication-terminator protein complexed with DNA., Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K, Nature. 1996 Oct 17;383(6601):598-603. PMID:8857533
Page seeded by OCA on Thu Feb 21 12:26:21 2008