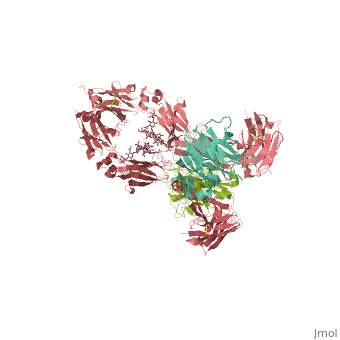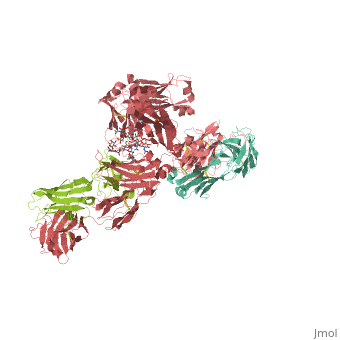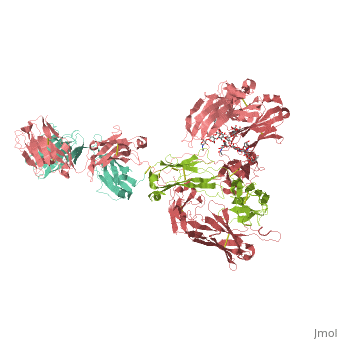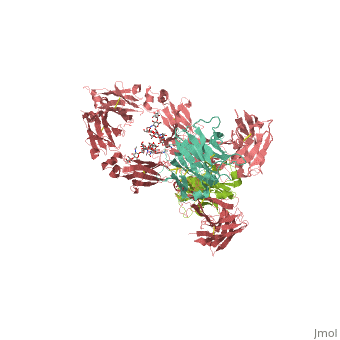We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
IgG Branco
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
== Your Heading Here (maybe something like 'Structure') == | == Your Heading Here (maybe something like 'Structure') == | ||
| - | <StructureSection load='1hzh' size='350' side='right' caption=' | + | <StructureSection load='1hzh' size='350' side='right' caption='Crystallographic structure of human IgG, with the Protein Data Bank (PDB) code 1HZH (PDB entry [[1hzh]])' scene=''> |
Affinity chromatography with protein A from Staphylococcus aureus (SpA) is the most widespread and accepted methodology for antibody capture during the downstream process of antibody manufacturing. A triazine based ligand (ligand 22/8) was previously developed as an inexpensive and robust alternative to SpA chromatography (Li et al. [12] and Teng et al. [11]). Despite the experimental success, there is no structural information on the binding modes of ligand 22/8 to antibodies, namely to Immunoglobulin G (IgG) molecules and fragments. In this work, we addressed this issue by a molecular docking approach allied to molecular dynamics simulations. Theoretical results confirmed the preference of the synthetic ligand to bind IgG through the binding site found in the crystallographic structure of the natural complex between SpA and the Fc fragment of IgG. Our studies also suggested other unknown “hot-spots” for specific binding of the affinity ligand at the hinge between VH and CH1 domains of Fab fragment. The best docking poses were further analysed by molecular dynamics studies at three different protonation states (pH 3, 7 and 11). The main interactions between ligand 22/8 and the IgG fragments found at pH 7 were weaker at pH 3 and pH 11 and in these conditions the ligand start losing tight contact with the binding site, corroborating the experimental evidence for protein elution from the chromatographic adsorbents at these pH conditions. | Affinity chromatography with protein A from Staphylococcus aureus (SpA) is the most widespread and accepted methodology for antibody capture during the downstream process of antibody manufacturing. A triazine based ligand (ligand 22/8) was previously developed as an inexpensive and robust alternative to SpA chromatography (Li et al. [12] and Teng et al. [11]). Despite the experimental success, there is no structural information on the binding modes of ligand 22/8 to antibodies, namely to Immunoglobulin G (IgG) molecules and fragments. In this work, we addressed this issue by a molecular docking approach allied to molecular dynamics simulations. Theoretical results confirmed the preference of the synthetic ligand to bind IgG through the binding site found in the crystallographic structure of the natural complex between SpA and the Fc fragment of IgG. Our studies also suggested other unknown “hot-spots” for specific binding of the affinity ligand at the hinge between VH and CH1 domains of Fab fragment. The best docking poses were further analysed by molecular dynamics studies at three different protonation states (pH 3, 7 and 11). The main interactions between ligand 22/8 and the IgG fragments found at pH 7 were weaker at pH 3 and pH 11 and in these conditions the ligand start losing tight contact with the binding site, corroborating the experimental evidence for protein elution from the chromatographic adsorbents at these pH conditions. | ||
| - | |||
</StructureSection> | </StructureSection> | ||
Revision as of 08:13, 6 September 2012
Your Heading Here (maybe something like 'Structure')
| |||||||||||