This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox christian
From Proteopedia
| Line 12: | Line 12: | ||
'''Crystall structure [[1tca]] of '''CALB''''''. | '''Crystall structure [[1tca]] of '''CALB''''''. | ||
| - | |||
| - | <imagemap> | ||
| - | Image:Image:1-s2 0-S1381117712001348-gr2.jpg|200px|picture of a foo | ||
| - | poly 131 45 213 41 210 110 127 109 [[2ace]] | ||
| - | poly 104 126 105 171 269 162 267 124 [[1rat]] | ||
| - | rect 15 95 94 176 [http://en.wikipedia.org/wiki/Student] | ||
| - | # A comment, this line is ignored | ||
| - | circle 57 57 20 [[Believe_It_or_Not!]] | ||
| - | desc bottom-left | ||
| - | </imagemap> | ||
| - | |||
| - | |||
| - | |||
| - | <imagemap> | ||
| - | Image:1-s20-S1381117712001348-gr2.jpg|250px|CALB | ||
| - | rect 238,217,303,243 [http://en.wikipedia.org/wiki/Student] | ||
| - | rect 344,224,399,240 [http://en.wikipedia.org/wiki/Student] | ||
| - | rect 403,223,459,239 [http://en.wikipedia.org/wiki/Student] | ||
| - | desc bottom-left | ||
| - | </imagemap> | ||
| - | |||
The <scene name='Sandbox_christian/Active_site/1'>Active Site</scene> consists of '''Ser 105''', '''Asp 187''' and '''His 224'''. Click here to see the <scene name='Sandbox_christian/Active_site/2'> B-factors</scene> and the <scene name='Sandbox_christian/Active_site/3'>Glycosylation site</scene>. | The <scene name='Sandbox_christian/Active_site/1'>Active Site</scene> consists of '''Ser 105''', '''Asp 187''' and '''His 224'''. Click here to see the <scene name='Sandbox_christian/Active_site/2'> B-factors</scene> and the <scene name='Sandbox_christian/Active_site/3'>Glycosylation site</scene>. | ||
Revision as of 11:21, 6 September 2012
Candida antarctica lipase B
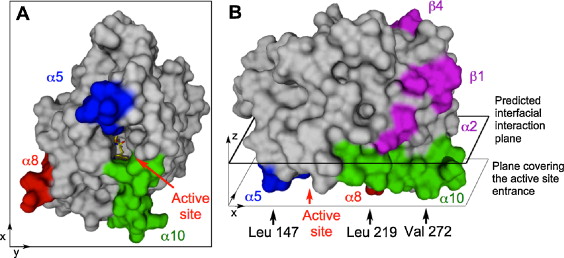
Lipase B from Candida antarctica binds to hydrophobic substrate–water interfaces via hydrophobic anchors surrounding the active site entrance [1]
Abstract: Candida antarctica Lipase B (CALB) has been extensively studied over the past decades and has proved to be an efficient catalyst for various industrial and scientific applications. Because of its ability to hydrolyse soluble and insoluble substrates and the lack of a classical interfacial activation it was previously characterized as an intermediate between a lipase and an esterase. We show by molecular dynamics simulation in full atomistic detail that CALB attaches and binds to a hydrophobic tributyrin–water interface via three hydrophobic anchor regions defined by Leu 147, Leu 219, and Val 272 surrounding the entrance to the active site. These regions trigger the reorientation of the protein via hydrophobic interactions even when the protein impacts at the surface in a non-optimal orientation. During the binding process the flexible helix α5 undergoes a movement of 7.5 Å towards the substrate layer. Though tributyrin has no net charge a long-range attraction between interface and protein was observed up to a separation of 7 Å that corresponds to the amplitude of the fluctuations of the tributyrin surface. A stable binding of the protein with the active site oriented towards the active site was observed for several hundreds of nanoseconds in total. During that time, single tributyrin molecules moved from the tributyrin layer into the substrate binding site of CALB and were temporarily binding as productive sn3-complexes.
| |||||||||||
Recently MD simulations showed that CALB binds to hydrophobic interfaces [2] and [3].
- ↑ C. Gruber, J. Pleiss: Lipase B from Candida antarctica binds to hydrophobic substrate-water interfaces via hydrophobic anchors surrounding the active site entrance, J MOL CATAL B-ENZYM, 2012; http://dx.doi.org/10.1016/j.molcatb.2012.05.012
- ↑ Gruber CC, Pleiss J. Molecular modeling of lipase binding to a substrate-water interface. Methods Mol Biol. 2012;861:313-27. PMID:22426727 doi:10.1007/978-1-61779-600-5_19
- ↑ Gruber CC, Pleiss J. Systematic benchmarking of large molecular dynamics simulations employing GROMACS on massive multiprocessing facilities. J Comput Chem. 2011 Mar;32(4):600-6. doi: 10.1002/jcc.21645. Epub 2010 Sep 1. PMID:20812321 doi:10.1002/jcc.21645