This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1js3
From Proteopedia
(New page: 200px<br /><applet load="1js3" size="450" color="white" frame="true" align="right" spinBox="true" caption="1js3, resolution 2.25Å" /> '''Crystal structure of...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1js3.gif|left|200px]]<br /><applet load="1js3" size=" | + | [[Image:1js3.gif|left|200px]]<br /><applet load="1js3" size="350" color="white" frame="true" align="right" spinBox="true" |
caption="1js3, resolution 2.25Å" /> | caption="1js3, resolution 2.25Å" /> | ||
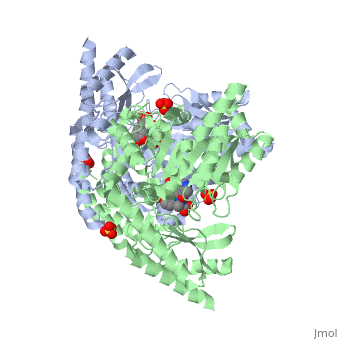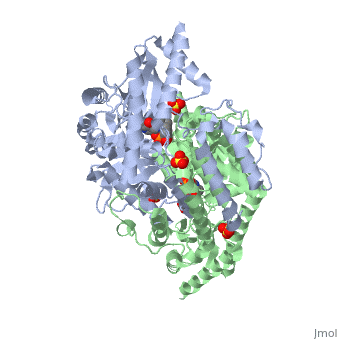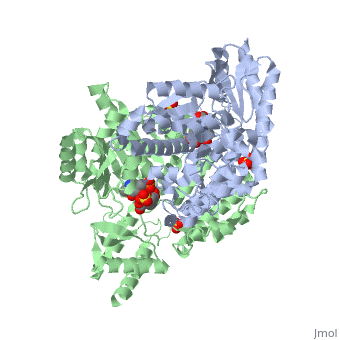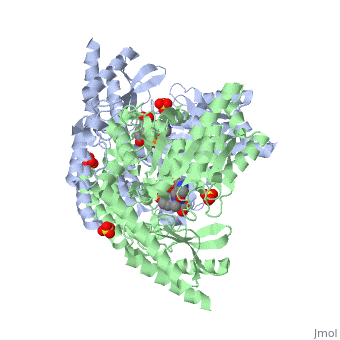
'''Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa'''<br /> | '''Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa'''<br /> | ||
==Overview== | ==Overview== | ||
| - | DOPA decarboxylase (DDC) is responsible for the synthesis of the key | + | DOPA decarboxylase (DDC) is responsible for the synthesis of the key neurotransmitters dopamine and serotonin via decarboxylation of L-3,4-dihydroxyphenylalanine (L-DOPA) and L-5-hydroxytryptophan, respectively. DDC has been implicated in a number of clinic disorders, including Parkinson's disease and hypertension. Peripheral inhibitors of DDC are currently used to treat these diseases. We present the crystal structures of ligand-free DDC and its complex with the anti-Parkinson drug carbiDOPA. The inhibitor is bound to the enzyme by forming a hydrazone linkage with the cofactor, and its catechol ring is deeply buried in the active site cleft. The structures provide the molecular basis for the development of new inhibitors of DDC with better pharmacological characteristics. |
==About this Structure== | ==About this Structure== | ||
| - | 1JS3 is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Sus_scrofa Sus scrofa] with SO4, PLP and 142 as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Aromatic-L-amino-acid_decarboxylase Aromatic-L-amino-acid decarboxylase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.1.28 4.1.1.28] Full crystallographic information is available from [http:// | + | 1JS3 is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Sus_scrofa Sus scrofa] with <scene name='pdbligand=SO4:'>SO4</scene>, <scene name='pdbligand=PLP:'>PLP</scene> and <scene name='pdbligand=142:'>142</scene> as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Aromatic-L-amino-acid_decarboxylase Aromatic-L-amino-acid decarboxylase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.1.28 4.1.1.28] Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JS3 OCA]. |
==Reference== | ==Reference== | ||
| Line 17: | Line 17: | ||
[[Category: Burkhard, P.]] | [[Category: Burkhard, P.]] | ||
[[Category: Dominici, P.]] | [[Category: Dominici, P.]] | ||
| - | [[Category: Jansonius, J | + | [[Category: Jansonius, J N.]] |
| - | [[Category: Malashkevich, V | + | [[Category: Malashkevich, V N.]] |
[[Category: 142]] | [[Category: 142]] | ||
[[Category: PLP]] | [[Category: PLP]] | ||
| Line 27: | Line 27: | ||
[[Category: vitamin b6]] | [[Category: vitamin b6]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 13:26:09 2008'' |
Revision as of 11:26, 21 February 2008
|
Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa
Overview
DOPA decarboxylase (DDC) is responsible for the synthesis of the key neurotransmitters dopamine and serotonin via decarboxylation of L-3,4-dihydroxyphenylalanine (L-DOPA) and L-5-hydroxytryptophan, respectively. DDC has been implicated in a number of clinic disorders, including Parkinson's disease and hypertension. Peripheral inhibitors of DDC are currently used to treat these diseases. We present the crystal structures of ligand-free DDC and its complex with the anti-Parkinson drug carbiDOPA. The inhibitor is bound to the enzyme by forming a hydrazone linkage with the cofactor, and its catechol ring is deeply buried in the active site cleft. The structures provide the molecular basis for the development of new inhibitors of DDC with better pharmacological characteristics.
About this Structure
1JS3 is a Single protein structure of sequence from Sus scrofa with , and as ligands. Active as Aromatic-L-amino-acid decarboxylase, with EC number 4.1.1.28 Full crystallographic information is available from OCA.
Reference
Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase., Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN, Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243
Page seeded by OCA on Thu Feb 21 13:26:09 2008