1kcg
From Proteopedia
(New page: 200px<br /> <applet load="1kcg" size="450" color="white" frame="true" align="right" spinBox="true" caption="1kcg, resolution 2.6Å" /> '''NKG2D in complex wit...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1kcg.gif|left|200px]]<br /> | + | [[Image:1kcg.gif|left|200px]]<br /><applet load="1kcg" size="350" color="white" frame="true" align="right" spinBox="true" |
| - | <applet load="1kcg" size=" | + | |
caption="1kcg, resolution 2.6Å" /> | caption="1kcg, resolution 2.6Å" /> | ||
'''NKG2D in complex with ULBP3'''<br /> | '''NKG2D in complex with ULBP3'''<br /> | ||
==Overview== | ==Overview== | ||
| - | NKG2D is known to trigger the natural killer (NK) cell lysis of various | + | NKG2D is known to trigger the natural killer (NK) cell lysis of various tumor and virally infected cells. In the NKG2D/ULBP3 complex, the structure of ULBP3 resembles the alpha1 and alpha2 domains of classical MHC molecules without a bound peptide. The lack of alpha3 and beta2m domains is compensated by replacing two hydrophobic patches at the underside of the class I MHC-like beta sheet floor with a group of hydrophilic and charged residues in ULBP3. NKG2D binds diagonally across the ULBP3 alpha helices, creating a complementary interface, an asymmetrical subunit orientation, and local conformational adjustments in the receptor. The interface is stabilized primarily by hydrogen bonds and hydrophobic interactions. Unlike the KIR receptors that recognize a conserved HLA region by a lock-and-key mechanism, NKG2D recognizes diverse ligands by an induced-fit mechanism. |
==About this Structure== | ==About this Structure== | ||
| - | 1KCG is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] with GSH as [http://en.wikipedia.org/wiki/ligand ligand]. Full crystallographic information is available from [http:// | + | 1KCG is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] with <scene name='pdbligand=GSH:'>GSH</scene> as [http://en.wikipedia.org/wiki/ligand ligand]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1KCG OCA]. |
==Reference== | ==Reference== | ||
| Line 21: | Line 20: | ||
[[Category: protein-protein complex]] | [[Category: protein-protein complex]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 13:32:30 2008'' |
Revision as of 11:32, 21 February 2008
|
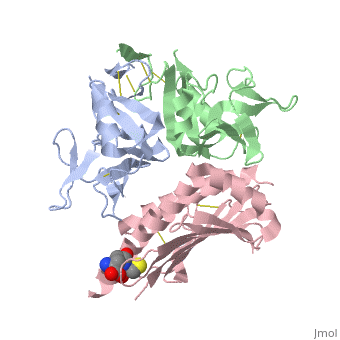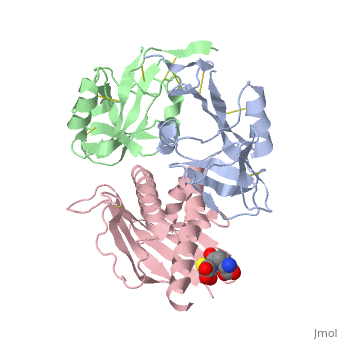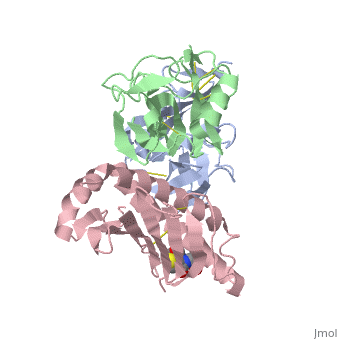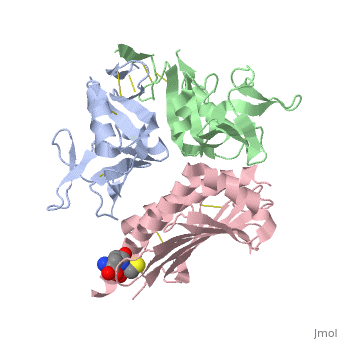
NKG2D in complex with ULBP3
Overview
NKG2D is known to trigger the natural killer (NK) cell lysis of various tumor and virally infected cells. In the NKG2D/ULBP3 complex, the structure of ULBP3 resembles the alpha1 and alpha2 domains of classical MHC molecules without a bound peptide. The lack of alpha3 and beta2m domains is compensated by replacing two hydrophobic patches at the underside of the class I MHC-like beta sheet floor with a group of hydrophilic and charged residues in ULBP3. NKG2D binds diagonally across the ULBP3 alpha helices, creating a complementary interface, an asymmetrical subunit orientation, and local conformational adjustments in the receptor. The interface is stabilized primarily by hydrogen bonds and hydrophobic interactions. Unlike the KIR receptors that recognize a conserved HLA region by a lock-and-key mechanism, NKG2D recognizes diverse ligands by an induced-fit mechanism.
About this Structure
1KCG is a Protein complex structure of sequences from Homo sapiens with as ligand. Full crystallographic information is available from OCA.
Reference
Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3., Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD, Immunity. 2001 Dec;15(6):1039-49. PMID:11754823
Page seeded by OCA on Thu Feb 21 13:32:30 2008