Journal:JBSD:30
From Proteopedia
(Difference between revisions)

| Line 25: | Line 25: | ||
'''4. Conformational heterogeneity observed in MD simulations''' | '''4. Conformational heterogeneity observed in MD simulations''' | ||
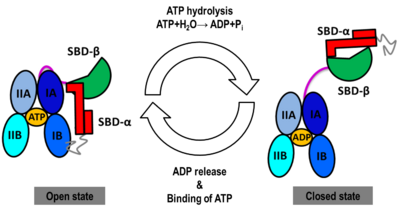
Prompted by the absence of detailed atomic experimental structures, we carried out the task of generating computer models of the human Hsp70 in its open and closed states by homology modeling and by molecular dynamics (MD) simulations on a unprecedented micro-second timescale, using the templates PDB ID: [[3c7n]] and PDB ID: [[2kho]], respectively. The sequence of human inducible Hsp70 (UniProt ID: P08107) was chosen for modeling purposes because it has been reported as a new target for cancer therapies. | Prompted by the absence of detailed atomic experimental structures, we carried out the task of generating computer models of the human Hsp70 in its open and closed states by homology modeling and by molecular dynamics (MD) simulations on a unprecedented micro-second timescale, using the templates PDB ID: [[3c7n]] and PDB ID: [[2kho]], respectively. The sequence of human inducible Hsp70 (UniProt ID: P08107) was chosen for modeling purposes because it has been reported as a new target for cancer therapies. | ||
| - | We obtained stable structures by using six and three different initial conditions in the molecular dynamics (MD) simulations of the open and closed models, respectively. In the six MD trajectories, the open model adopted a stable conformation in which the lid was open, the SBD-α was docked to the NBD (Figure 2) and the linker was bound to a cleft between subdomains IA and IIA of the NBD. Different stable binding positions sites were found for the SBD-α on the lobe I of the NBD (Figure 2). The conformational heterogeneity of the closed model, probed by three MD trajectories corresponded to a rotation of the SBD around the NBD, in agreement with NMR data (Figure 2). | + | We obtained stable structures by using six and three different initial conditions in the molecular dynamics (MD) simulations of the open and closed models, respectively. In the six MD trajectories, the <scene name='Journal:JBSD:30/Cv/11'>open model</scene> adopted a stable conformation in which the lid was open, the SBD-α was docked to the NBD (Figure 2) and the linker was bound to a cleft between subdomains IA and IIA of the NBD. Different stable binding positions sites were found for the SBD-α on the lobe I of the NBD (Figure 2). The conformational heterogeneity of the closed model, probed by three MD trajectories corresponded to a rotation of the SBD around the NBD, in agreement with NMR data (Figure 2). |
We found that human Hsp70, both in its open and closed states, was better represented by an ensemble of conformations on a μs timescale, corresponding to different local minima of its free-energy landscape in good agreement with low-resolution experimental data of Hsp70s homologs (Small Angle Xray-Scattering and single-molecule Förster Resonance Energy Transfer) to which the MD simulations were compared in details. These structural ensembles (Figure 2) represent the first attempt to model the conformational heterogeneity of a full-length molecular chaperone at the atomic scale. | We found that human Hsp70, both in its open and closed states, was better represented by an ensemble of conformations on a μs timescale, corresponding to different local minima of its free-energy landscape in good agreement with low-resolution experimental data of Hsp70s homologs (Small Angle Xray-Scattering and single-molecule Förster Resonance Energy Transfer) to which the MD simulations were compared in details. These structural ensembles (Figure 2) represent the first attempt to model the conformational heterogeneity of a full-length molecular chaperone at the atomic scale. | ||
Revision as of 10:56, 3 October 2012
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.