1p7q
From Proteopedia
| Line 4: | Line 4: | ||
==Overview== | ==Overview== | ||
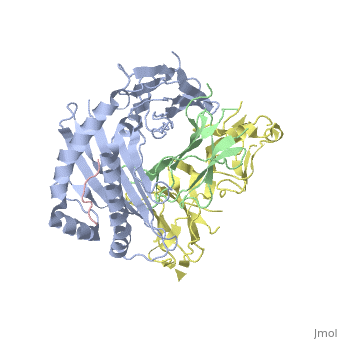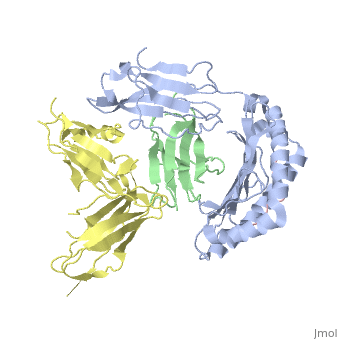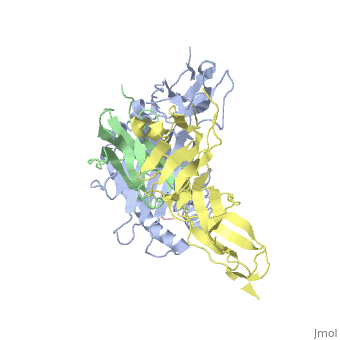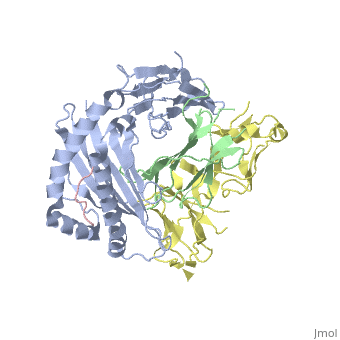
| - | Leukocyte immunoglobulin-like receptor 1 (LIR-1), an inhibitory receptor | + | Leukocyte immunoglobulin-like receptor 1 (LIR-1), an inhibitory receptor expressed on monocytes, dendritic cells and lymphocytes, regulates cellular function by binding a broad range of classical and nonclassical major histocompatibility complex (MHC) class I molecules, and the human cytomegalovirus MHC class I homolog UL18. Here we describe the 3.4-A crystal structure of a complex between the LIR-1 D1D2 domains and the MHC class I molecule HLA-A2. LIR-1 contacts the mostly conserved beta(2)-microglobulin and alpha3 domains of HLA-A2. The LIR-1 binding site comprises residues at the interdomain hinge, and a patch at the D1 tip. The structure shows how LIR-1 recognizes UL18 and diverse MHC class I molecules, and indicates that a similar mode of MHC class I recognition is used by other LIR family members. |
==Disease== | ==Disease== | ||
| Line 16: | Line 16: | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
| - | [[Category: Bjorkman, P | + | [[Category: Bjorkman, P J.]] |
| - | [[Category: Thomas, L | + | [[Category: Thomas, L M.]] |
| - | [[Category: Willcox, B | + | [[Category: Willcox, B E.]] |
[[Category: hla-a2/lir-1 complex]] | [[Category: hla-a2/lir-1 complex]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 14:26:11 2008'' |
Revision as of 12:26, 21 February 2008
|
Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor
Contents |
Overview
Leukocyte immunoglobulin-like receptor 1 (LIR-1), an inhibitory receptor expressed on monocytes, dendritic cells and lymphocytes, regulates cellular function by binding a broad range of classical and nonclassical major histocompatibility complex (MHC) class I molecules, and the human cytomegalovirus MHC class I homolog UL18. Here we describe the 3.4-A crystal structure of a complex between the LIR-1 D1D2 domains and the MHC class I molecule HLA-A2. LIR-1 contacts the mostly conserved beta(2)-microglobulin and alpha3 domains of HLA-A2. The LIR-1 binding site comprises residues at the interdomain hinge, and a patch at the D1 tip. The structure shows how LIR-1 recognizes UL18 and diverse MHC class I molecules, and indicates that a similar mode of MHC class I recognition is used by other LIR family members.
Disease
Known diseases associated with this structure: Abacavir hypersensitivity, susceptibility to OMIM:[142800], Ankylosing spondylitis, susceptibility to, 1 OMIM:[142800], Hypoproteinemia, hypercatabolic OMIM:[109700], Stevens-Johnson syndrome, susceptibility to OMIM:[142800]
About this Structure
1P7Q is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor., Willcox BE, Thomas LM, Bjorkman PJ, Nat Immunol. 2003 Sep;4(9):913-9. Epub 2003 Aug 3. PMID:12897781
Page seeded by OCA on Thu Feb 21 14:26:11 2008