We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Grb10 SH2 Domain
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
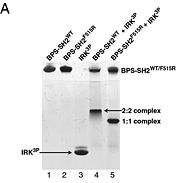
To ensure the crystallographic structure of Grb10 SH2 is indeed a dimer in solution, Evan Stein and colleagues substituted Phe515 at the dimer interface with arginine (electrically charged side chain) and found, using gel filtration chromatography, that the Grb10 SH2 dimer had indeed become independent monomers. | To ensure the crystallographic structure of Grb10 SH2 is indeed a dimer in solution, Evan Stein and colleagues substituted Phe515 at the dimer interface with arginine (electrically charged side chain) and found, using gel filtration chromatography, that the Grb10 SH2 dimer had indeed become independent monomers. | ||
| - | + | [[Image:Figure_6.jpeg | thumb | alt=text | asdflj]] | |
| + | |||
</StructureSection> | </StructureSection> | ||
Revision as of 05:52, 8 November 2012
| |||||||||||
Interaction Between Grb10 and E3 Ubiquitin Ligase NEDD4
WOOOOOOOO
|
Grb10 Gene Inhibition Affects Body Composition, and Insulin Signaling
References
- ↑ 1.0 1.1 1.2 Stein EG, Ghirlando R, Hubbard SR. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J Biol Chem. 2003 Apr 11;278(15):13257-64. Epub 2003 Jan 27. PMID:12551896 doi:http://dx.doi.org/10.1074/jbc.M212026200