We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Grb10 SH2 Domain
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==Dimerization of the Grb10 SH2 Domain== | ==Dimerization of the Grb10 SH2 Domain== | ||
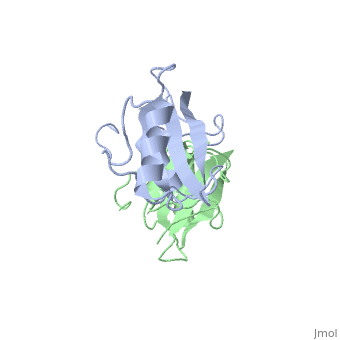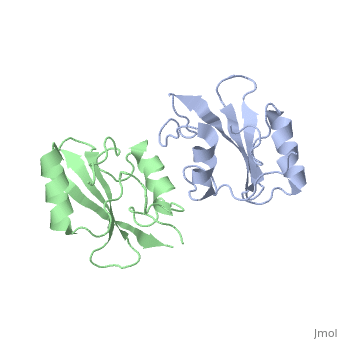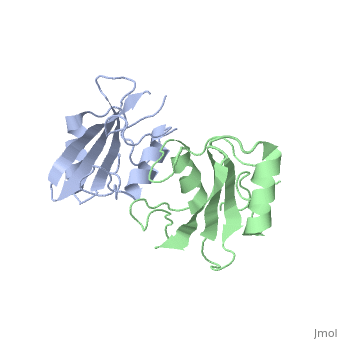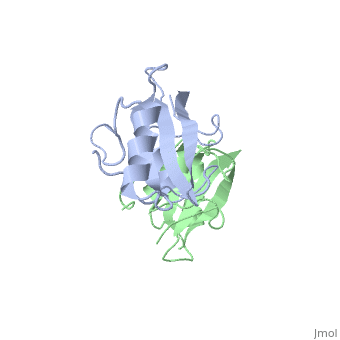
| - | The crystal structure of Grb10 SH2 domain (molecular mass = 12.4 kDa) was an important step to understanding how this protein interacts with IGF1 receptors, and although the SH2 domain functions as an independent segment, it forms a dimer in physiological environments. The <scene name='Grb10_SH2_Domain/Best_interface/1'>dimer interface</scene> exists due to the middle hydrophobic Phe515 and uncharged Thr504 (labeled blue) residues packed into its equivalent counter parter on the other protomer, designated as Phe515' and Thr504' (labeled red); Gln511 (blue) forms a hydrogen bond to the backbone of Asp514' (red) while the side chain of Asn519 forms two hydrogen bonds to the backbone of Lys505'. <ref name=Guan>PMID: 12551896 </ref> | + | The crystal structure of Grb10 SH2 domain (molecular mass = 12.4 kDa) was an important step to understanding how this protein interacts with IGF1 receptors, and although the SH2 domain functions as an independent segment, it forms a dimer in physiological environments. <ref name=Guan>PMID: 12551896 </ref> The <scene name='Grb10_SH2_Domain/Best_interface/1'>dimer interface</scene> exists due to the middle hydrophobic Phe515 and uncharged Thr504 (labeled blue) residues packed into its equivalent counter parter on the other protomer, designated as Phe515' and Thr504' (labeled red); Gln511 (blue) forms a hydrogen bond to the backbone of Asp514' (red) while the side chain of Asn519 forms two hydrogen bonds to the backbone of Lys505'. <ref name=Guan>PMID: 12551896 </ref> The interface ends with Leu518 and Phe-496' via hydrophobic interactions. <ref name=Guan>PMID: 12551896 </ref>. The structure of Grb10 SH2 forms similar SH2 domains found in other proteins, which have an <scene name='Grb10_SH2_Domain/Alpha_helix/1'>alpha helix</scene> on the outsides with anti-parallel <scene name='Grb10_SH2_Domain/Beta_sheet/1'>beta sheets</scene>. |
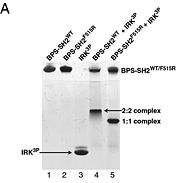
To ensure the crystallographic structure of Grb10 SH2 is indeed a dimer in solution, Evan Stein and colleagues substituted Phe515 at the dimer interface with arginine (electrically charged side chain) and found, using gel filtration chromatography (Picture 1), that the Grb10 SH2 dimer had indeed become independent monomers. | To ensure the crystallographic structure of Grb10 SH2 is indeed a dimer in solution, Evan Stein and colleagues substituted Phe515 at the dimer interface with arginine (electrically charged side chain) and found, using gel filtration chromatography (Picture 1), that the Grb10 SH2 dimer had indeed become independent monomers. | ||
| Line 13: | Line 13: | ||
[[Image:Figure_6.jpeg | thumb | alt=text | Picture 1]] | [[Image:Figure_6.jpeg | thumb | alt=text | Picture 1]] | ||
| + | ====Picture 1==== | ||
| + | |||
| + | The gel used for filtration chromatography had 5 lanes with 5 different components: | ||
| + | # BPS-SH2_WT (Wild Type) | ||
| + | # BPS-SH2_F515R (mutant = Phe515 --> Arg) | ||
| + | # IRK_3P | ||
| + | # BPS-SH2_WT + IRK_3P | ||
| + | # BPS-SH2_F515R + IRK_3P | ||
| + | |||
| + | As seen in lanes 1 and 2, the BPS-SH2 proteins did not travel down the gel due to their high pI; to resolve this issue, the researchers added IRK_3P to the two BPS-SH2 proteins which then made a complex that was mobile. <ref name=Guan>PMID: 12551896 </ref> Lane 4 shows a band labeled ''2:2 complex'' that shows the position of the SH2 dimer. The additional band found at the very top of lane 4 represents the BPS-SH2_WT protein that did not complex with high motility protein IRK_3P, i.e. it was not able to migrate through the gel due to its high pI. Lane 5 shows a band labeled ''1:1 complex'' elucidating that the Arg substitution at Phe515 did indeed produce a monomer, which was able to travel farther down the gel. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 06:29, 8 November 2012
| |||||||||||
Interaction Between Grb10 and E3 Ubiquitin Ligase NEDD4
WOOOOOOOO
|