We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
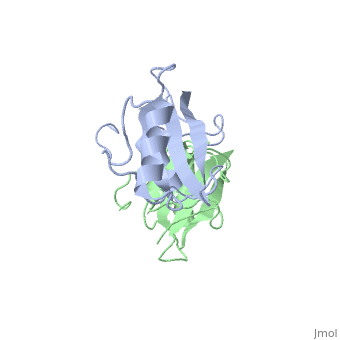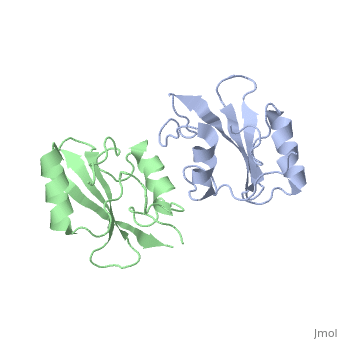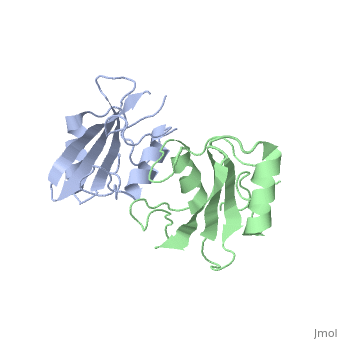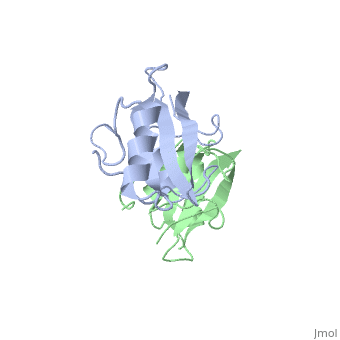
Grb10 SH2 Domain
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
# BPS-SH2_WT (Wild Type) | # BPS-SH2_WT (Wild Type) | ||
# BPS-SH2_F515R (mutant = Phe515 --> Arg) | # BPS-SH2_F515R (mutant = Phe515 --> Arg) | ||
| - | # IRK_3P | + | # IRK_3P (tris-phosphorylated Insulin Receptor Kinase domain) |
# BPS-SH2_WT + IRK_3P | # BPS-SH2_WT + IRK_3P | ||
# BPS-SH2_F515R + IRK_3P | # BPS-SH2_F515R + IRK_3P | ||
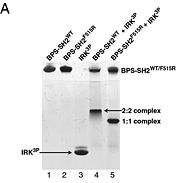
As seen in lanes 1 and 2, the BPS-SH2 proteins did not travel down the gel due to their high pI; to resolve this issue, the researchers added IRK_3P to the two BPS-SH2 proteins which then made a complex that was mobile. <ref name=Guan>PMID: 12551896 </ref> Lane 4 shows a band labeled ''2:2 complex'' that shows the position of the SH2 dimer. The additional band found at the very top of lane 4 represents the BPS-SH2_WT protein that did not complex with high motility protein IRK_3P, i.e. it was not able to migrate through the gel due to its high pI. Lane 5 shows a band labeled ''1:1 complex'' elucidating that the Arg substitution at Phe515 did indeed produce a monomer, which was able to travel farther down the gel. | As seen in lanes 1 and 2, the BPS-SH2 proteins did not travel down the gel due to their high pI; to resolve this issue, the researchers added IRK_3P to the two BPS-SH2 proteins which then made a complex that was mobile. <ref name=Guan>PMID: 12551896 </ref> Lane 4 shows a band labeled ''2:2 complex'' that shows the position of the SH2 dimer. The additional band found at the very top of lane 4 represents the BPS-SH2_WT protein that did not complex with high motility protein IRK_3P, i.e. it was not able to migrate through the gel due to its high pI. Lane 5 shows a band labeled ''1:1 complex'' elucidating that the Arg substitution at Phe515 did indeed produce a monomer, which was able to travel farther down the gel. | ||
| + | |||
| + | '''Why BPS-SH2?''' | ||
| + | |||
| + | In order for the full-length Grb10 protein to interact with Insulin Receptor Kinase (IRK), the SH2 and BPS domain must be present. <ref name=Guan>PMID: 9506989 </ref> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 07:26, 8 November 2012
| |||||||||||
Interaction Between Grb10 and E3 Ubiquitin Ligase NEDD4
WOOOOOOOO
|