Grb10 SH2 Domain
From Proteopedia
(→There are 3 interfaces at which Nedd4 C2 and Grb10 SH2 interact:) |
(→There are 3 interfaces at which Nedd4 C2 and Grb10 SH2 interact:) |
||
| Line 44: | Line 44: | ||
| - | <scene name='Grb10_SH2_Domain/Interface_iii/1'>Interface III</scene> is made of NEDD4 C2's proline rich C terminus which complexes with Grb10 H2 N-terminal residues 431-440 and C-terminal residues 532-535; among this interaction <scene name='Grb10_SH2_Domain/Interface_iii_hbond/1'>ARG533 (SH2) and PRO284 (C2)</scene> forms a hydrogen bond and <scene name='Grb10_SH2_Domain/Interface_iii_saltbridge/1'>ARG431 (SH2) and PRO287 (C2)</scene> forms a slat bridge | + | <scene name='Grb10_SH2_Domain/Interface_iii/1'>Interface III</scene> is made of NEDD4 C2's proline rich C terminus which complexes with Grb10 H2 N-terminal residues 431-440 and C-terminal residues 532-535; among this interaction <scene name='Grb10_SH2_Domain/Interface_iii_hbond/1'>ARG533 (SH2) and PRO284 (C2)</scene> forms a hydrogen bond and <scene name='Grb10_SH2_Domain/Interface_iii_saltbridge/1'>ARG431 (SH2) and PRO287 (C2)</scene> forms a slat bridge. <ref>PMID: 20980250</ref> |
==Grb10 Gene Inhibition Affects Body Composition, and Insulin Signaling== | ==Grb10 Gene Inhibition Affects Body Composition, and Insulin Signaling== | ||
Revision as of 12:24, 8 November 2012
| |||||||||||
Contents |
Interaction Between Grb10 and E3 Ubiquitin Ligase NEDD4
|
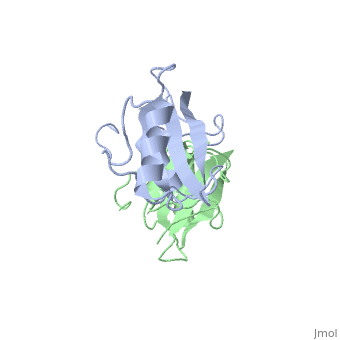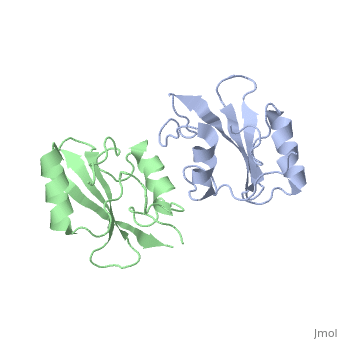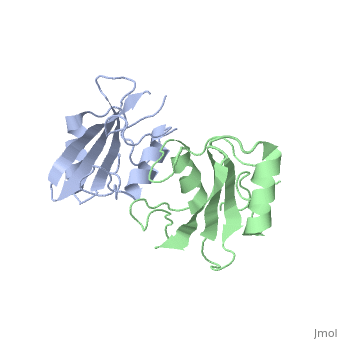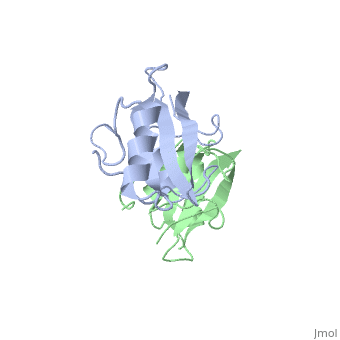
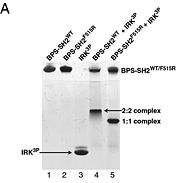
Grb10 has now been shown to not only inhibit insulin receptors and IGF1R kinase activity, but also interacts via its SH2 domain with the C2 domain of E3 ubiquitin ligase NEDD4 facilitating ubiquitation of IGF1R [3]. It is hypothesized that the Grb10 SH2 interaction with the E3 domain of NEDD4 may allow NEDD4 to come in close proximity to IGF1R promoting degradation.[4]
There are 3 interfaces at which Nedd4 C2 and Grb10 SH2 interact:
is the largest of the 3 interfaces between Grb10 SH2 and NEDD4 C2 domains. There are 8 hydrogen bonds formed at this interface, with the least important H bonds formed between , for disruption between these two residues with an induced mutation did not inhibit Grb10 SH2 and NEDD4 C2 to complex. [5]
, the smallest of the 3, is composed of residues 429-434 of Grb10 SH2 interacting with residues 112-114 of NEDD4 C2; disruption of this interaction does not disrupt the SH2-C2 domain complex. [6]
is made of NEDD4 C2's proline rich C terminus which complexes with Grb10 H2 N-terminal residues 431-440 and C-terminal residues 532-535; among this interaction forms a hydrogen bond and forms a slat bridge. [7]
Grb10 Gene Inhibition Affects Body Composition, and Insulin Signaling
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Stein EG, Ghirlando R, Hubbard SR. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J Biol Chem. 2003 Apr 11;278(15):13257-64. Epub 2003 Jan 27. PMID:12551896 doi:http://dx.doi.org/10.1074/jbc.M212026200
- ↑ He W, Rose DW, Olefsky JM, Gustafson TA. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem. 1998 Mar 20;273(12):6860-7. PMID:9506989
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412