Sandbox Reserved 646
From Proteopedia
| Line 16: | Line 16: | ||
== '''Structure''' == | == '''Structure''' == | ||
| - | ''' | + | '''Isoforms & Topology'' |
| + | Arginase is unique because of all the enzymes found in the urea cycle this enzyme has different isoforms. In the human body there are two isozymes with similar structural properties and differing immunological properties. Common among the differing isozymes is the requirement of two divalent cation cofactors for activity. Manganese2+ is the most common cofactor for activity but cobalt, nickel and iron have been reported to be suitable cofactors as well. The two required manganese II ions are located in two separate subunits and are separated by approximately 3.36-3.57 Å and bridged by oxygen and a solvent derived hydroxide <ref>Ash, David Structure and Function of Arginases [http://www.ncbi.nlm.nih.gov/pubmed/15465781]</ref> Many varying isoforms of arginase have produced different structural models based on the particular isoform and organism of origin. The most well studied arginase enzyme, arginase I isolated from rat liver, is a trimeric enzyme and each subunit is ordered in an alpha-beta-alpha structure. Each fold contains an eight-stranded, parallel β-sheet surrounded by alpha helices, with an approximate size 35kDa <ref>Ash, David Structure and Function of Arginases [http://www.ncbi.nlm.nih.gov/pubmed/15465781]</ref>. Fully active enzymes contain a binuclear metal cofactor center in a 15Å cleft that serves as the active site. One cofactor is held in place by terminal ligands His 101 and Asp 128, and bridged by Asp 124, Asp 132 and hydroxide ion <ref>Ash, David Structure and Function of Arginases [http://www.ncbi.nlm.nih.gov/pubmed/15465781]</ref>. The other cofactor held by terminal ligand residues His 101 and Asp 234 with bridging ligands Asp 232 and hydroxide ion <ref>Ash, David Structure and Function of Arginases [http://www.ncbi.nlm.nih.gov/pubmed/15465781]</ref>. | ||
| + | |||
| + | Although arginase is present in different isoforms, there is a high level of conservation among the amino acids. For example, human type I and IIarginases share a 58% sequence identity (Ash). Overall, there are 20 amino acids that are highly conserved over the varying isoforms. Most of these conserved residues are key structural residues. Included in these conserved structural residues is gly 23, which begins the first α-helix of arginase(Perozich). In addition to highly conserved nucleotides, there exist a few key invariant nucleotides as well. Among the most notable are Val 120, Ile 121, Val 233 and Ile 264 which are attributed to the packing of the hydrophobic interior of the protein (Perozich). | ||
| + | |||
| + | |||
| - | '''Topology''' | ||
== '''Medical Application''' == | == '''Medical Application''' == | ||
Revision as of 06:10, 15 November 2012
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
Arginase
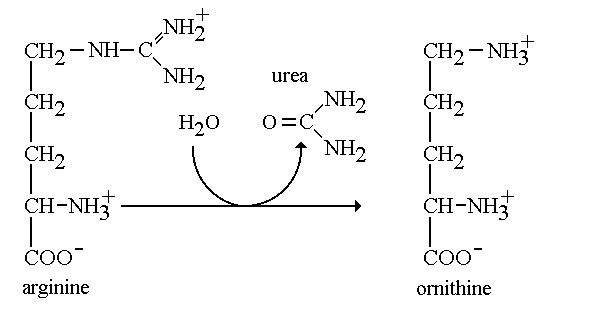
Arginase is an enzyme found within the important urea cycle. Arginase is the final enzyme that allows the Urea cycle to complete its fifth and last step. Arginase takes on two distinct forms: Class I and II. The grouping into classes refers to the catalytic activity that the enzyme is capable of performing. In addition, each class are encoded by a different gene. Arginase I (arg1) is found in the cytoplasm of liver cells functioning in the urea cycle, while arginase II (arg2) is found in the mitochondria and functions in the kidney. In interest of our studies, we will refer to arginase I as arginase. Arginase belongs to the enzymatic class hydrolase. Arginase functions by cleaving the amino acid arginine with water to produce urea. When one studies enzymes, he pays careful attention the it’s specific activity. Specific activity quantifies the amount of product formed by the enzyme in reference to a given time per concentration of a protein. Arginase is known to have high specific activity and functions to produce a good amount of urea.[1] In addition, this enzyme allows the regeneration of ornithine which is the final product of the urea cycle. When there is a deficiency in the arg1 gene it is understood that one is undergoing an urea cycle disorder. In people with arginase deficiency, arginase is either missing or harmed, therefore arginine is not metabolized correctly. In lieu of this, it is impossible for urea to be formed, leaving the excess nitrogen to accumulate in the blood in the form of ammonia. Which leads to serious problems in the body.[2] MechanismArginine + H2O → Ornithine + Urea The mechanism regarding arginase is highly important in the production of urea through the urea cycle. During the final step of the urea cycle, the amino acid arginine is present and needs to be cleaved in order for urea to be produced. Arginine is hydrolyzed and cleaved with the hydroxyl at the end of the amino group. We refer to this group as a guanidium end. The guanidium group is positively charged and has a high pKa. Therefore, it is willing to donate a proton since it has a hydrogen to give. First, the ligand will ionize the water to form hydroxide. The hydroxide now attacks the guanidine carbon and protons are transferred from the hydroxide to the substrate bridging. In the third and final step protons are transferred through a proton shuffle to create ornithine and urea. Afterwards, the urea is excreted through urine. Leaving the excess ornithine to be recycled through the final step in the cycle to react with citrulline and eliminate ammonia from the body. [3] Structure'Isoforms & Topology Arginase is unique because of all the enzymes found in the urea cycle this enzyme has different isoforms. In the human body there are two isozymes with similar structural properties and differing immunological properties. Common among the differing isozymes is the requirement of two divalent cation cofactors for activity. Manganese2+ is the most common cofactor for activity but cobalt, nickel and iron have been reported to be suitable cofactors as well. The two required manganese II ions are located in two separate subunits and are separated by approximately 3.36-3.57 Å and bridged by oxygen and a solvent derived hydroxide [4] Many varying isoforms of arginase have produced different structural models based on the particular isoform and organism of origin. The most well studied arginase enzyme, arginase I isolated from rat liver, is a trimeric enzyme and each subunit is ordered in an alpha-beta-alpha structure. Each fold contains an eight-stranded, parallel β-sheet surrounded by alpha helices, with an approximate size 35kDa [5]. Fully active enzymes contain a binuclear metal cofactor center in a 15Å cleft that serves as the active site. One cofactor is held in place by terminal ligands His 101 and Asp 128, and bridged by Asp 124, Asp 132 and hydroxide ion [6]. The other cofactor held by terminal ligand residues His 101 and Asp 234 with bridging ligands Asp 232 and hydroxide ion [7]. Although arginase is present in different isoforms, there is a high level of conservation among the amino acids. For example, human type I and IIarginases share a 58% sequence identity (Ash). Overall, there are 20 amino acids that are highly conserved over the varying isoforms. Most of these conserved residues are key structural residues. Included in these conserved structural residues is gly 23, which begins the first α-helix of arginase(Perozich). In addition to highly conserved nucleotides, there exist a few key invariant nucleotides as well. Among the most notable are Val 120, Ile 121, Val 233 and Ile 264 which are attributed to the packing of the hydrophobic interior of the protein (Perozich).
Medical ApplicationThe urea cycle is important for removing urea from the body so that the body is able to regulate and carry on its normal functions. However, if the body is not able to get dispose of urea, it spells trouble for specific bodily functions. Currently, we understand that arginase functions to break down the amino acid arginine into ornithine while releasing urea. However, we know that the deficiency of arginase is strictly inherited and thereby a genetic disorder. When argininase is deficient, it causes a phenomenon of a build up in ammonia and arginine. An accumulation of high levels of ammonia can be toxic to the body and causes neurological effects.
SymptomsSpastic tetraplegia Progressive mental retardation Seizures Hyperactivity Growth failure Elevated blood ammonia level Enlarged liver Lack of appetite Vomiting Increased blood level of arginine Muscle stiffness Spasticity Tremor Balance problems Coordination problems Irritability |