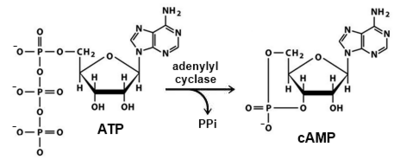
Adenylyl cyclase
From Proteopedia
| Line 30: | Line 30: | ||
Although adenylyl cyclase is found throughout organisms at a universal level, distantly related organisms have different modifications of the enzyme, each is specialized for a particular task in a particular environment<ref name="Tews">PMID:15890882</ref>. As stated earlier humans have 10 known isozymes of adenylyl cyclase; whereas ''Escherichia coli'' has only one isozyme, and ''Mycobacterium tuberculosis'' has 15<ref name="Tews"/>. One particularly interesting adenylyl cyclase possessed by ''M. tuberculosis'', Rv1264, has a N-terminal which, in a sense, acts as a pH sensor, as it regulates the activity of the enzyme based on the pH of the surrounding solution<ref name="Linder"/><ref name="Tews"/>. This adenylyl cyclase, like most others, belongs to class III, adenylyl cyclases in this class have multiple domains, at least one for catalysis, and another for regulation<ref name="Tews"/>. | Although adenylyl cyclase is found throughout organisms at a universal level, distantly related organisms have different modifications of the enzyme, each is specialized for a particular task in a particular environment<ref name="Tews">PMID:15890882</ref>. As stated earlier humans have 10 known isozymes of adenylyl cyclase; whereas ''Escherichia coli'' has only one isozyme, and ''Mycobacterium tuberculosis'' has 15<ref name="Tews"/>. One particularly interesting adenylyl cyclase possessed by ''M. tuberculosis'', Rv1264, has a N-terminal which, in a sense, acts as a pH sensor, as it regulates the activity of the enzyme based on the pH of the surrounding solution<ref name="Linder"/><ref name="Tews"/>. This adenylyl cyclase, like most others, belongs to class III, adenylyl cyclases in this class have multiple domains, at least one for catalysis, and another for regulation<ref name="Tews"/>. | ||
| - | <StructureSection load='1y11' size='450' frame='true' align='right' caption='Rv1264 adenylyl cyclase monomer in its active state.' scene='Sandbox_159/Main_1y11/2'> | ||
=== Structure === | === Structure === | ||
| - | [[Image:Dimer2.png|300px|left]]The <scene name='Adenylyl_cyclase/Main_1y11/1'>Rv1264</scene> adenylyl cyclase is a 363 residue long protein composed of a catalytic <scene name='Adenylyl_cyclase/Cdomain_1y11/1'>C-terminal domain</scene> and a regulatory <scene name='Adenylyl_cyclase/Ndomain_1y11/1'>N-terminal domain</scene> which contains a flexible <scene name='Adenylyl_cyclase/Connector_1y11/1'>linker region</scene> which connects the two<ref name="Tews"/>. The active structure is a homodimer resembling the mammalian type II homodimer, where an α-helix of one monomer (Chain A) is placed through the central coiled coil of another (Chain B)<ref name="Zhang"/><ref name="Tews"/>. This dimerization places the regulatory domain of chain A in close proximity to the catalytic domain of chain B, and vice versa<ref name="Tews"/>. Two switch elements are present in the protein, one in the C-terminal domain (α1-switch) and another in the linker region (αN10-switch), these allow for large conformational changes to take place in the enzyme in response to relatively small environmental changes<ref name="Linder"/><ref name="Tews"/>. | + | [[Image:Dimer2.png|300px|left]] |
| + | The <scene name='Adenylyl_cyclase/Main_1y11/1'>Rv1264</scene> adenylyl cyclase is a 363 residue long protein composed of a catalytic <scene name='Adenylyl_cyclase/Cdomain_1y11/1'>C-terminal domain</scene> and a regulatory <scene name='Adenylyl_cyclase/Ndomain_1y11/1'>N-terminal domain</scene> which contains a flexible <scene name='Adenylyl_cyclase/Connector_1y11/1'>linker region</scene> which connects the two<ref name="Tews"/>. The active structure is a homodimer resembling the mammalian type II homodimer, where an α-helix of one monomer (Chain A) is placed through the central coiled coil of another (Chain B)<ref name="Zhang"/><ref name="Tews"/>. This dimerization places the regulatory domain of chain A in close proximity to the catalytic domain of chain B, and vice versa<ref name="Tews"/>. Two switch elements are present in the protein, one in the C-terminal domain (α1-switch) and another in the linker region (αN10-switch), these allow for large conformational changes to take place in the enzyme in response to relatively small environmental changes<ref name="Linder"/><ref name="Tews"/>. | ||
==== C-Terminal Catalytic Domain ==== | ==== C-Terminal Catalytic Domain ==== | ||
The catalytic activity in Rv1264's <scene name='Adenylyl_cyclase/Active_1y11/1'>active site</scene> is performed by the residues: Asp 222 (Red), Lys 261 (Blue), Asp 265 (Orange), Arg 298 (Pink), Asp 312 (Yellow), Asn 319 (Purple), Arg 323 (Green)<ref name="Tews"/>. All of these residues create a highly polar environment which is complementary in charge and polarity to the intermediate of the reaction<ref name="Linder"/><ref name="Tews"/>. Residues which guide the phosphates of ATP, arginine 298 and 323, bind a <scene name='Adenylyl_cyclase/Sulphate_1y11/1'>sulphate ion</scene> in the active site<ref name="Tesmer2">PMID:10427002</ref>. This sulphate ion is located in the position which will be occupied by the ATP's β-phosphate during catalysis<ref name="Tesmer2"/>. In the process of catalysis the β-phosphate is cleaved from the α-phosphate; this reaction may be made more favourable by lowering the energy though complementary charge associations between the β-phosphate and arginines 298 and 323<ref name="Tews"/><ref name="Tesmer2"/>. Another residue, <scene name='Adenylyl_cyclase/Argglycerol_1y11/1'>arginine 296</scene>, binds a <scene name='Adenylyl_cyclase/Glycerol_1y11/1'>glycerol</scene> molecule through electrostatic interactions<ref name="Tews"/>. The specific function of this association is unknown, but because of its close proximity to the active site it may play a role in catalysis<ref name="Tews"/>. | The catalytic activity in Rv1264's <scene name='Adenylyl_cyclase/Active_1y11/1'>active site</scene> is performed by the residues: Asp 222 (Red), Lys 261 (Blue), Asp 265 (Orange), Arg 298 (Pink), Asp 312 (Yellow), Asn 319 (Purple), Arg 323 (Green)<ref name="Tews"/>. All of these residues create a highly polar environment which is complementary in charge and polarity to the intermediate of the reaction<ref name="Linder"/><ref name="Tews"/>. Residues which guide the phosphates of ATP, arginine 298 and 323, bind a <scene name='Adenylyl_cyclase/Sulphate_1y11/1'>sulphate ion</scene> in the active site<ref name="Tesmer2">PMID:10427002</ref>. This sulphate ion is located in the position which will be occupied by the ATP's β-phosphate during catalysis<ref name="Tesmer2"/>. In the process of catalysis the β-phosphate is cleaved from the α-phosphate; this reaction may be made more favourable by lowering the energy though complementary charge associations between the β-phosphate and arginines 298 and 323<ref name="Tews"/><ref name="Tesmer2"/>. Another residue, <scene name='Adenylyl_cyclase/Argglycerol_1y11/1'>arginine 296</scene>, binds a <scene name='Adenylyl_cyclase/Glycerol_1y11/1'>glycerol</scene> molecule through electrostatic interactions<ref name="Tews"/>. The specific function of this association is unknown, but because of its close proximity to the active site it may play a role in catalysis<ref name="Tews"/>. | ||
Revision as of 13:11, 15 November 2012
| |||||||||||
Contents |
Biological Role
M. tuberculosis is a pathogenic bacterium, and thus it faces an array of a host's immune responses to attempt in an attempt to rid of it[20]. One of the hosts defense mechanisms M. tuberculosis faces is acidification encountered in phagolysosomes. The ability to be able to detect this acidic environment, and have an appropriate response to it may greatly assist M. tuberculosis infect a host[21][22]. As cAMP levels are increased, acidification of other structures is delayed and elevated cAMP levels activate cAMP receptor proteins which in turn regulate transcription[23][24].
3D Structures of Adenylyl cyclase
2ev1, 2ev2, 2ev3, 2ev4 – MtADCY N-terminal – Mycobacterium tuberculosis
1y10, 1y11 – MtADCY holoenzyme
2fjt – ADCY 4 – Yersinia pestis
1ykd – ADCY – Anabaena
1ab8 – ADCY C2 domain - rat
ADCY catalytic domain
1yk9, 1ybt, 1ybu – MtADCY catalytic domain
1fx2, 1fx4 - ADCY catalytic domain – Trypanosoma brucei
2bw7 - SpADCY catalytic domain+catechol estrogen – Spirulina platensis
1wc0 – SpADCY catalytic domain+methylene ATP
1wc1, 1wc3, 1wc4, 1wc5, 1wc6 - SpADCY catalytic domain+ATP derivatives
3maa – dADCY 5 (mutant)+ADCY 2 C2a+guanine nucleotide binding protein G – dog
3g82 - dADCY 5 (mutant)+ADCY 2 C1a+guanine nucleotide binding protein G
3c14, 3c15 - dADCY 5 C1a (mutant)+ADCY 2 C2a+guanine nucleotide binding protein G+PPi+ions
3c16 - dADCY 5 C1a (mutant)+ADCY 2 C2a+guanine nucleotide binding protein G+ATP+Ca
2gvd, 2gvz - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+ATP+Mn
1tl7, 1u0h - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GTP+Mn
1cs4 - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+AMP+Mg+PPI+GDP
1cul - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GDP+Mg+triphosphate
1cjk - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+ATP+Mn+Mg
1cjt - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GDP+Mn+Mg+ATP
1cju - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GDP+Mg+ATP
1cjv - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GDP+Mg+Zn+ATP
1azs - dADCY 5 C1a +ADCY 2 C2a+guanine nucleotide binding protein G+GDP+Mg
1yrt, 1yru - ADCY catalytic domain+calmodulin C-terminal – Bordetella pertussis
ADCY anthrax edema factor associated domain
1k8t - BaADCY C-terminal - Bacillus anthracis
1lvc, 1pk0, 1s26 – BaADCY C-terminal+calmodulin+anthranyloyl ATP+Yb+Ca
1sk6 - BaADCY C-terminal+calmodulin+cAMP+PPi
1xfu - BaADCY C-terminal (mutant)+calmodulin
1xfv, 1k90 - BaADCY C-terminal+calmodulin+dATP
1xfw - BaADCY C-terminal+calmodulin+cAMP
1xfx, 1xfz - BaADCY C-terminal+calmodulin+Ca
1xfy, 1k93 - BaADCY C-terminal+calmodulin
1y0v - BaADCY C-terminal+calmodulin+PPi
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Taussig R, Gilman AG. Mammalian membrane-bound adenylyl cyclases. J Biol Chem. 1995 Jan 6;270(1):1-4. PMID:7814360
- ↑ 2.0 2.1 2.2 2.3 2.4 Hurley JH. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J Biol Chem. 1999 Mar 19;274(12):7599-602. PMID:10075642
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Zhang G, Liu Y, Ruoho AE, Hurley JH. Structure of the adenylyl cyclase catalytic core. Nature. 1997 Mar 20;386(6622):247-53. PMID:9069282 doi:10.1038/386247a0
- ↑ Ketkar AD, Shenoy AR, Kesavulu MM, Visweswariah SS, Suguna K. Purification, crystallization and preliminary X-ray diffraction analysis of the catalytic domain of adenylyl cyclase Rv1625c from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2004 Feb;60(Pt 2):371-3. Epub 2004, Jan 23. PMID:14747729 doi:10.1107/S0907444903028002
- ↑ Beis I, Newsholme EA. The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem J. 1975 Oct;152(1):23-32. PMID:1212224
- ↑ Haring HU, Renner R, Hepp KD. Hormonal control of cyclic AMP turnover in isolated fat cells. Mol Cell Endocrinol. 1976 Aug-Sep;5(3-4):295-302. PMID:182581
- ↑ Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003 Dec 5;278(49):49523-9. Epub 2003 Sep 25. PMID:14512417 doi:10.1074/jbc.M309543200
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 8.8 8.9 Linder JU, Schultz A, Schultz JE. Adenylyl cyclase Rv1264 from Mycobacterium tuberculosis has an autoinhibitory N-terminal domain. J Biol Chem. 2002 May 3;277(18):15271-6. Epub 2002 Feb 11. PMID:11839758 doi:10.1074/jbc.M200235200
- ↑ 9.0 9.1 9.2 Siddappa R, Martens A, Doorn J, Leusink A, Olivo C, Licht R, van Rijn L, Gaspar C, Fodde R, Janssen F, van Blitterswijk C, de Boer J. cAMP/PKA pathway activation in human mesenchymal stem cells in vitro results in robust bone formation in vivo. Proc Natl Acad Sci U S A. 2008 May 20;105(20):7281-6. Epub 2008 May 19. PMID:18490653
- ↑ Cox RP, Gilbert P Jr, Griffin MJ. Alkaline inorganic pyrophosphatase activity of mammalian-cell alkaline phosphatase. Biochem J. 1967 Oct;105(1):155-61. PMID:4964763
- ↑ Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717-49. PMID:9242922 doi:10.1146/annurev.biochem.66.1.717
- ↑ 12.0 12.1 Feinstein PG, Schrader KA, Bakalyar HA, Tang WJ, Krupinski J, Gilman AG, Reed RR. Molecular cloning and characterization of a Ca2+/calmodulin-insensitive adenylyl cyclase from rat brain. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10173-7. PMID:1719547
- ↑ 13.0 13.1 13.2 13.3 13.4 Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997 Dec 12;278(5345):1907-16. PMID:9417641
- ↑ Masters SB, Sullivan KA, Miller RT, Beiderman B, Lopez NG, Ramachandran J, Bourne HR. Carboxyl terminal domain of Gs alpha specifies coupling of receptors to stimulation of adenylyl cyclase. Science. 1988 Jul 22;241(4864):448-51. PMID:2899356
- ↑ 15.0 15.1 15.2 15.3 15.4 Wang M, Ramos BP, Paspalas CD, Shu Y, Simen A, Duque A, Vijayraghavan S, Brennan A, Dudley A, Nou E, Mazer JA, McCormick DA, Arnsten AF. Alpha2A-adrenoceptors strengthen working memory networks by inhibiting cAMP-HCN channel signaling in prefrontal cortex. Cell. 2007 Apr 20;129(2):397-410. PMID:17448997 doi:10.1016/j.cell.2007.03.015
- ↑ 16.00 16.01 16.02 16.03 16.04 16.05 16.06 16.07 16.08 16.09 16.10 16.11 16.12 16.13 16.14 16.15 16.16 16.17 16.18 16.19 16.20 16.21 16.22 16.23 16.24 16.25 16.26 16.27 16.28 16.29 16.30 16.31 16.32 16.33 16.34 16.35 16.36 Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, Linder JU. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science. 2005 May 13;308(5724):1020-3. PMID:15890882 doi:http://dx.doi.org/308/5724/1020
- ↑ 17.0 17.1 17.2 Tesmer JJ, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, Sprang SR. Two-metal-Ion catalysis in adenylyl cyclase. Science. 1999 Jul 30;285(5428):756-60. PMID:10427002
- ↑ Tesmer JJ, Dessauer CW, Sunahara RK, Murray LD, Johnson RA, Gilman AG, Sprang SR. Molecular basis for P-site inhibition of adenylyl cyclase. Biochemistry. 2000 Nov 28;39(47):14464-71. PMID:11087399
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 19.7 19.8 Findeisen F, Linder JU, Schultz A, Schultz JE, Brugger B, Wieland F, Sinning I, Tews I. The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid. J Mol Biol. 2007 Jun 22;369(5):1282-95. Epub 2007 Apr 12. PMID:17482646 doi:10.1016/j.jmb.2007.04.013
- ↑ Glickman MS, Jacobs WR Jr. Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell. 2001 Feb 23;104(4):477-85. PMID:11239406
- ↑ Channon JY, Kasper LH. Parasite subversion of the host cell endocytic network. Parasitol Today. 1995 Feb;11(2):47-8. PMID:15275372
- ↑ Warner DF, Mizrahi V. The survival kit of Mycobacterium tuberculosis. Nat Med. 2007 Mar;13(3):282-4. PMID:17342138 doi:10.1038/nm0307-282
- ↑ Lowrie DB, Jackett PS, Ratcliffe NA. Mycobacterium microti may protect itself from intracellular destruction by releasing cyclic AMP into phagosomes. Nature. 1975 Apr 17;254(5501):600-2. PMID:165421
- ↑ Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, Whalan R, Hinds J, Colston MJ, Green J, Buxton RS. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005 Jun;56(5):1274-86. PMID:15882420 doi:10.1111/j.1365-2958.2005.04609.x
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Michal Harel, David Canner, Basavraj Khanppnavar, Travis Eyford, Joel L. Sussman