Sandbox Reserved 390
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
==COMPOUND ACTIVE SITE== | ==COMPOUND ACTIVE SITE== | ||
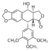
| - | This molecule has 36 <scene name='Sandbox_Reserved_390/Top/3'>alpha helices</scene> represented with magenta helices and 40 <scene name='Sandbox_Reserved_390/Top/2'>beta sheets</scene> represented with blue arrows in the <scene name='Sandbox_Reserved_390/Top/4'>secondary structures</scene>. At the active site we can see how the <scene name='Sandbox_Reserved_390/Top/14'>ligand</scene> (in green) is stabilized. The detail view of the <scene name='Sandbox_Reserved_390/New/5'>active site</scene> show how the ligand is stabilized by amino acid <scene name='Sandbox_Reserved_390/Arg/1'>arginine</scene> (at position 503B in blue), <scene name='Sandbox_Reserved_390/New/4'>aspartic acid </scene> (at position 479B in yellow with hydrogen bond interaction), <scene name='Sandbox_Reserved_390/Gln/1'>glutamine</scene> (at position 778B in purple), also by nucleoside <scene name='Sandbox_Reserved_390/New/7'>thymidine</scene>(in light blue at position 9F) and <scene name='Sandbox_Reserved_390/New/6'>deoxyguanosine</scene> (in green at possition 13D showing a Pi-Pi interaction) | + | This molecule has 36 <scene name='Sandbox_Reserved_390/Top/3'>alpha helices</scene> represented with magenta helices and 40 <scene name='Sandbox_Reserved_390/Top/2'>beta sheets</scene> represented with blue arrows in the <scene name='Sandbox_Reserved_390/Top/4'>secondary structures</scene>. At the active site we can see how the <scene name='Sandbox_Reserved_390/Top/14'>ligand</scene> (in green) is stabilized. The detail view of the <scene name='Sandbox_Reserved_390/New/5'>active site</scene> show how the ligand is stabilized by amino acid <scene name='Sandbox_Reserved_390/Arg/1'>arginine</scene> (at position 503B in blue), <scene name='Sandbox_Reserved_390/New/4'>aspartic acid </scene> (at position 479B in yellow with hydrogen bond interaction), <scene name='Sandbox_Reserved_390/Gln/1'>glutamine</scene> (at position 778B in purple), also by nucleoside <scene name='Sandbox_Reserved_390/New/7'>thymidine</scene> (in light blue at position 9F) and <scene name='Sandbox_Reserved_390/New/6'>deoxyguanosine</scene> (in green at possition 13D showing a Pi-Pi interaction) |
==ETOPOSIDE RESISTANCE== | ==ETOPOSIDE RESISTANCE== | ||
Revision as of 00:27, 20 November 2012
Human topoisomerase IIbeta in complex with DNA and etoposide
| |||||||||||
References
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID:21778401 doi:10.1126/science.1204117
- ↑ Kathryn L. Gilroy, Chrysoula Leontiou, Kay Padget, Jeremy H. Lakey and Caroline A. Austin* "mAMSA resistant human topoisomerase IIβ mutation G465D has reduced ATP hydrolysis activity” Oxford JournalsLife Sciences Nucleic Acids Research Volume 34, Issue 5Pp. 1597-1607. DOI: 10.1093/nar/gkl057
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID:21778401 doi:10.1126/science.1204117