Tutorial:Basic Chemistry Topics
From Proteopedia
| Line 86: | Line 86: | ||
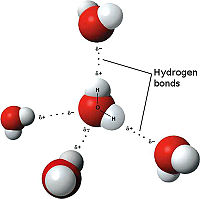
[[Image:3D hydrogen Bonding.jpg | thumb | left | 200px | Hydrogen Bonding<ref>Maňas, Michal, trans. "File:3D model hydrogen bonds in water.jpg." Wikimedia Commons. Wikimedia Commons, 3 Dec. 2007. Web. 31 Oct. 2012 <http://commons.wikimedia.org/wiki/File:3D_model_hydrogen_bonds_in_water.jpg.</ref>]] | [[Image:3D hydrogen Bonding.jpg | thumb | left | 200px | Hydrogen Bonding<ref>Maňas, Michal, trans. "File:3D model hydrogen bonds in water.jpg." Wikimedia Commons. Wikimedia Commons, 3 Dec. 2007. Web. 31 Oct. 2012 <http://commons.wikimedia.org/wiki/File:3D_model_hydrogen_bonds_in_water.jpg.</ref>]] | ||
| - | The weakest bond, the hydrogen bond is an attractive interaction between an electronegative atom and hydrogen. Electronegative atoms have high electron density. | + | The weakest bond, the hydrogen bond is an attractive interaction between an electronegative atom and hydrogen. Electronegative atoms have high electron density. High electron density refers to strong atoms that pull electrons towards it from weaker/low electron density atoms, such as hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. A common example of this is water. The image to the right demonstrates the hydrogen bonding in water. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons. When oxygen pulls the electrons, it leaves hydrogen with a slight positive charge. Since oxygen is pulling the hydrogen’s inward, the formation of a water droplet is possible. In this representation the hydrogen bonds are represented as yellow-dashed lines. The hydrogen bonds are important in this study and this molecular compound because they offer the stability of the secondary structures. <scene name='Tutorial:Basic_Chemistry_Topics/Hydrogen_bonds/2'>Hydrogen Bonds</scene> |
| Line 96: | Line 96: | ||
='''Primary Structure'''= | ='''Primary Structure'''= | ||
| + | The primary sequence of a compound is the amino acid linear sequence of the compound. For example the linear sequence of tobramycin is displayed below. The chemical formula is C18H37N5O9. If you count each of the elements the number should match. | ||
='''Secondary Structures'''= | ='''Secondary Structures'''= | ||
| - | Secondary structures are alpha helices and beta sheets. The helices and | + | Secondary structures are alpha helices and beta sheets. The helices and sheets provide stability to the molecule as a whole. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. Alpha helices have a cylinder-like structure that rotates in a clockwise manner with a parallel formation. This representation shows these key features of alpha helices. Here, you can see the helices rotating clockwise (follow the arrows), and the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. Beta sheets are often anti-parallel, which are clearly represented in this figure. The folding of a protein, alpha helices and beta sheets, is what gives the compound its function. When there is a change in protein folding, the function will change. |
<scene name='Tutorial:Basic_Chemistry_Topics/Alpha_beta_2ndstructures/1'>Alpha Helices and Beta Sheets</scene> | <scene name='Tutorial:Basic_Chemistry_Topics/Alpha_beta_2ndstructures/1'>Alpha Helices and Beta Sheets</scene> | ||
| Line 112: | Line 113: | ||
='''Active Site'''= | ='''Active Site'''= | ||
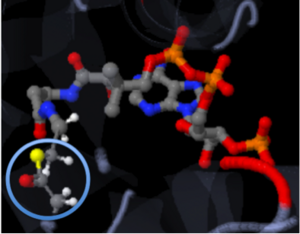
| - | The active site of a molecule can be described as a pocket where an interaction between compounds occurs. This interaction will cause a change in structure shape. The conformational change, change in structure shape, can inhibit or activate the physiological/pathological affect. The active site can either be inhibited or activated by the compound that binds there. Referring back to our article, the active site is where the acetylation is going to occur. In this depiction of the active site you can see the | + | The active site of a molecule can be described as a pocket where an interaction between compounds occurs. This interaction will cause a change in structure shape. The conformational change, change in structure shape, can inhibit or activate the physiological/pathological affect. The active site can either be inhibited or activated by the compound that binds there. Referring back to our article, the active site is where the acetylation is going to occur. In this depiction of the active site you can see the pocket where Coenzyme A (CoA) will aid the enzyme in the acetylation of Tobramycin (Toy), the aminoglycoside antibiotic. The enzyme is the "blob" surrounding the antibiotic and CoA. The enzyme/protein is holding these compounds in a conformation forcing them to react. The acetylation at the active site will cause the antibiotic to be inactive, hence inhibiting the active site. When Tobramycin becomes inactivated it is no longer able to aid in the destruction of bacteria. This is what we call antibiotic resistance. |
<scene name='Tutorial:Basic_Chemistry_Topics/Active_site/2'>Active Site</scene> | <scene name='Tutorial:Basic_Chemistry_Topics/Active_site/2'>Active Site</scene> | ||
Revision as of 16:32, 26 November 2012
Purpose of the Tutorial
- This tutorial is intended as a beneficial learning/teaching aid for an entry-level chemistry college student with some basic chemistry knowledge. This tutorial is based on learning, comprehending and applying their knowledge. Applying general chemistry to a research article will allow the students to see the impact they can have on the research world in the future by applying their knowledge. Various general chemistry topics are discussed in detail for an entry-level college student and then shown through various interactive representations of the compound studied by the research article.
Summary: Scientific Research Article
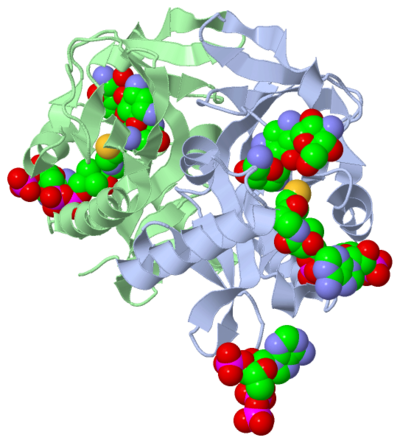
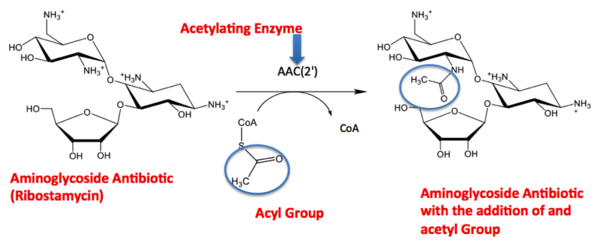
The molecule to left is from the article "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin" published in Nature Structural Biology. [2]. The study focused on aminoglycoside 2’- N- acetyltransferase (AAC (2’)- Ic), an enzyme. An enzyme is a compound that speeds the rate of a reaction.
The scientists involved in the study determined the structure of AAC (2’)-Ic from Mycobacterium tuberculosis, a pathogen. A pathogen is microorganism that can cause harm to the human body. The specific structure/protein fold of AAC (2’)-Ic, is placed in the GCN5-related N-acetyltransferase (GNAT) superfamily. The GNAT superfamily is a group of enzymes that are similar in structure. The protein fold is important because it determines the function of a compound. Since the GNAT superfamily and AAC(2’)-Ic have similar structures, they also have similar functions.[2]
Although the physiological function of AAC(2’)-Ic is not certain, the structure determined by the scientists allowed them to hypothesize the function. The AAC(2’)-Ic enzyme is located within the mycothiol (a component of the pathogen) structure. AAC(2’)-Ic may be capable of acetylating the aminoglycoside antibiotic. An acetylation is the addition of CH3CO onto a compound, which in this case is the antibiotic. When this occurs the aminoglycoside antibiotic becomes inactive. The basis of this study is important because when pathogens become resistant or inactive to commonly used antibiotics, an infection that used to be easily cured can now become severe and life threatening.[[2]
| |||||||||||
References
- ↑ Vetting, M. W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin." RCSB Protien DataBase. N.p., 28 Aug.2002. Web. 13 July 2011. http://www.rcsb.org/pdb/explore/explore.do?structureId=1M4D
- ↑ 2.0 2.1 2.2 2.3 2.4 Vetting, Matthew W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin."Nature Structural Biology 9.9 (2002): 653-58. Print.
- ↑ User:Cepheus. "Periodic Table." Wikipedia. N.p., 26 Feb. 2007. Web. 26 Nov. 2012. <http://en.wikipedia.org/wiki/File:Periodic_table.svg>.
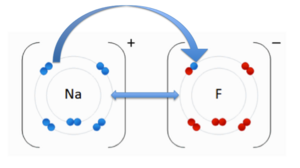
- ↑ . "File:NaF.gif." Wikipedia. Wikipedia, 17 June 2011. Web. 31 Oct. 2012.<http://en.wikipedia.org/wiki/File:NaF.gif.
- ↑ Maňas, Michal, trans. "File:3D model hydrogen bonds in water.jpg." Wikimedia Commons. Wikimedia Commons, 3 Dec. 2007. Web. 31 Oct. 2012 <http://commons.wikimedia.org/wiki/File:3D_model_hydrogen_bonds_in_water.jpg.
- ↑ 6.0 6.1 6.2 Wikipedia. Wikipedia, 4 Nov. 2012. Web. 7 Nov. 2012. <http://en.wikipedia.org/wiki/Enzyme_substrate_(biology)