We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Fragment-Based Drug Discovery
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
---- | ---- | ||
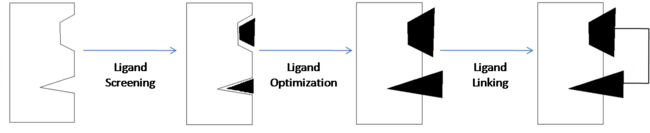
| - | ''' | + | One technique used in drug discovery is '''fragment-based drug discovery''' (FBDD). FBDD is a method of discovering new compounds by utilizing fragments that have some degree of binding affinity for a drug target, optimizing those fragments so as to increase their binding affinity, then linking them together to form a lead compound that has high affinity and selectivity for the drug target. Nuclear magnetic resonance (NMR) and x-ray crystallography can be used to analyze the fragments and drug targets in order to create three-dimensional images which can be used to obtain an analysis of molecular relationships. This allows developers to get a visual representation of how each fragment binds to the target and can also be useful in identifying the individual binding sites of the target. |
[[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery (Adapted from Fig. 1)<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref>]] | [[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery (Adapted from Fig. 1)<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref>]] | ||
| Line 17: | Line 17: | ||
! scope="col" width="5000px" | SAR by NMR | ! scope="col" width="5000px" | SAR by NMR | ||
|- | |- | ||
| - | | scope="col" width="5000px" | Structure-activity relationship (SAR) by NMR is one tool that | + | | scope="col" width="5000px" | Structure-activity relationship (SAR) by NMR is one tool that can be used to design and develop new drugs. This is the process "in which small organic molecules that bind to proximal subsites of a protein are identified, optimized, and linked together to produce high-affinity ligands."<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref> In other words, NMR is used to identify the components responsible for binding and analyze the relationship between the ligand and the biological target. |
|} | |} | ||
| Line 26: | Line 26: | ||
===== ABT-737: ligand screening ===== | ===== ABT-737: ligand screening ===== | ||
| - | <scene name='Sandbox_reserved_394/Compound_1/7'>Two fragments</scene> were found to have moderate affinity for Bcl-xl. <scene name='Sandbox_reserved_394/Compound_1/9'>Compound 1</scene> is a fluorobiphenylcarboxylic acid. It occupies <scene name='Sandbox_reserved_394/Binding_site_1/2'>binding site 1</scene> of Bcl-xl which consists of Phe 101, Tyr 105, Ala 108, Phe 109, Leu 136, Gly 142, Arg 143, and Ala 146. The fluorobiphenyl system of compound 1 is very hydrophobic and therefore, these residues form a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxylic acid portion of compound 1 binds near Gly 142</scene> of Bcl-xl. | + | <scene name='Sandbox_reserved_394/Compound_1/7'>Two fragments</scene> were found to have moderate affinity for Bcl-xl. <scene name='Sandbox_reserved_394/Compound_1/9'>Compound 1</scene> is a fluorobiphenylcarboxylic acid. It occupies <scene name='Sandbox_reserved_394/Binding_site_1/2'>binding site 1</scene> of Bcl-xl which consists of Phe 101, Tyr 105, Ala 108, Phe 109, Leu 136, Gly 142, Arg 143, and Ala 146. The fluorobiphenyl system of compound 1 is very hydrophobic and therefore, these residues form a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the system. There is also one hydrophilic interaction involved in this complex. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxylic acid portion of compound 1 binds near Gly 142</scene> of Bcl-xl. This is not a strong interaction but is significant because it can be modified to form a much stronger bond. |
| - | <scene name='Sandbox_reserved_394/Compound_1/3'>Compound 2</scene> is a napthalene-based alcohol which occupies <scene name='Sandbox_reserved_394/Binding_site_2/4'>binding site 2</scene> | + | <scene name='Sandbox_reserved_394/Compound_1/3'>Compound 2</scene> is a napthalene-based alcohol which occupies <scene name='Sandbox_reserved_394/Binding_site_2/4'>binding site 2</scene>. This particular fragment also is involved with hydrophobic interactions with Bcl-xl, although they are not as strong as in the case of compound 1. This binding site includes Ala 97, Glu 100, Phe 101, Val 145, and Tyr 199. |
==== Ligand Optimization ==== | ==== Ligand Optimization ==== | ||
| Line 51: | Line 51: | ||
|} | |} | ||
| - | + | One challenge in drug delivery is bioavailability. The bioavailibility may be decreased due to non-specific protein binding. Therefore, compound 3 required further optimization because the binding affinity for Bcl-xl is greatly reduced in the presence of human serum albumin (HSA). In order to decrease HSA affinity, and therefore increase Bcl-xl affinity, SAR by NMR was used to modify compound 3 by eliminating key binding groups of the compound to HSA without affecting Bcl-xl affinity. | |
{| class="wikitable collapsible" | {| class="wikitable collapsible" | ||
Revision as of 23:11, 27 November 2012
Drug Design: Fragment-Based Drug Discovery
| |||||||||||
References
- ↑ 1.0 1.1 Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.
- ↑ Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579
- ↑ Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf