Malarial Dihydrofolate Reductase as Drug Target
From Proteopedia
| Line 7: | Line 7: | ||
The | The | ||
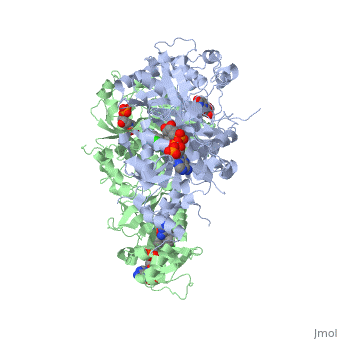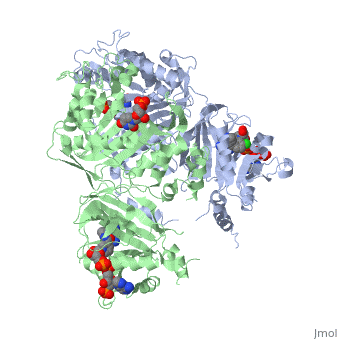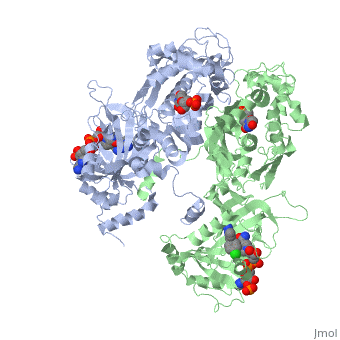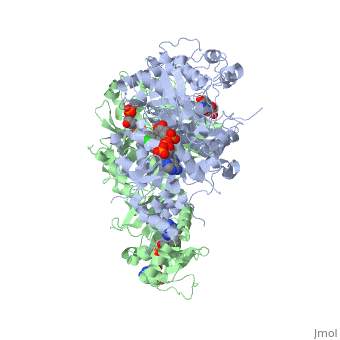
<scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Quadruple_mutant/2'>quadruple mutant</scene> of PfDHFR has four | <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Quadruple_mutant/2'>quadruple mutant</scene> of PfDHFR has four | ||
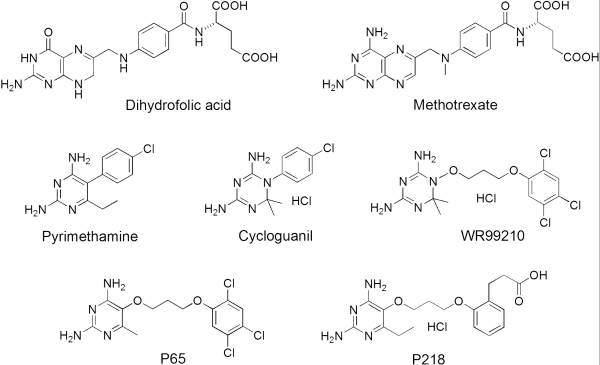
| - | <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Mutated_codons/2'>point mutations</scene>within the active site: N51I, C59R, S108N, and I164L.<ref>Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012 Jul 28;11:243. PMID: 22839209</ref> These mutations cause preferential decreased binding affinities of inhibitors, a huge factor being steric clashing, while having less of an effect on biological substrates. For pyrimethamine and cycloguanil specifically, the S108N mutation greatly effects their ability to bind the enzyme due to the drugs' rigid chlorophenyl substituents. The S108N mutation is also recognized as the the first mutation to have come about in the enzyme. The other mutations further decrease the binding affinities of inhibitors by resulting in conformational and other changes. In an effort to mitigate the effect of the mutations, the commonly occurring mutations and quadruple mutant must be addressed in the research of new PfDHFR targeting drugs. | + | <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Mutated_codons/2'>point mutations</scene> within the active site: N51I, C59R, S108N, and I164L.<ref>Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012 Jul 28;11:243. PMID: 22839209</ref> These mutations cause preferential decreased binding affinities of inhibitors, a huge factor being steric clashing, while having less of an effect on biological substrates. For pyrimethamine and cycloguanil specifically, the S108N mutation greatly effects their ability to bind the enzyme due to the drugs' rigid chlorophenyl substituents. The S108N mutation is also recognized as the the first mutation to have come about in the enzyme. The other mutations further decrease the binding affinities of inhibitors by resulting in conformational and other changes. In an effort to mitigate the effect of the mutations, the commonly occurring mutations and quadruple mutant must be addressed in the research of new PfDHFR targeting drugs. |
| Line 25: | Line 25: | ||
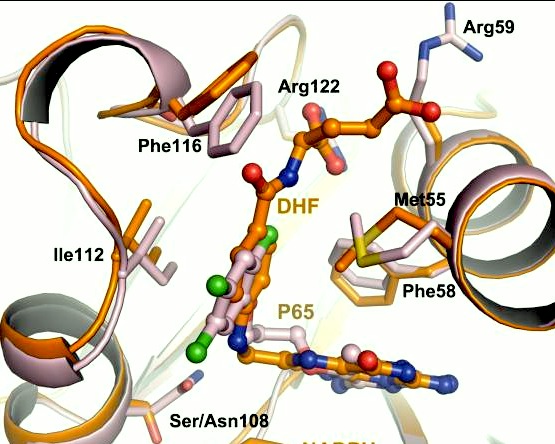
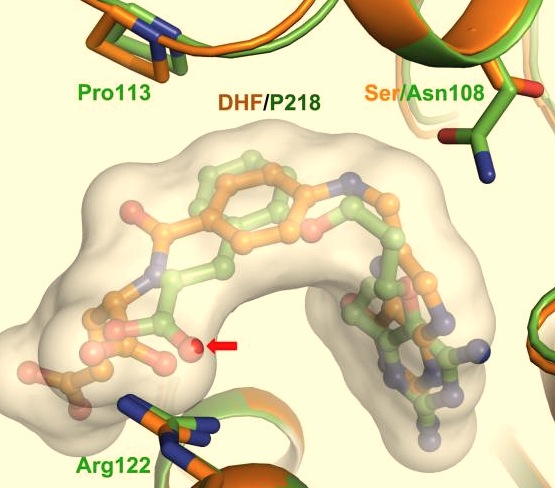
It was resolved that a structure containing 2,4-diaminopyrimidine anchor on a new drug would provide a solution for the steric hindrance present with pyrimethamine because of its rigid chlorophenyl group. In addition to this anchor allowing deep binding into the active site of PfDHFR, the other end of the molecule connected by a flexible linker region, a carboxylate group, would form strong hydrogen bonds with the conserved <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122/1'>Arg122</scene> residue. These ideas culminated into the formation of P218. | It was resolved that a structure containing 2,4-diaminopyrimidine anchor on a new drug would provide a solution for the steric hindrance present with pyrimethamine because of its rigid chlorophenyl group. In addition to this anchor allowing deep binding into the active site of PfDHFR, the other end of the molecule connected by a flexible linker region, a carboxylate group, would form strong hydrogen bonds with the conserved <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122/1'>Arg122</scene> residue. These ideas culminated into the formation of P218. | ||
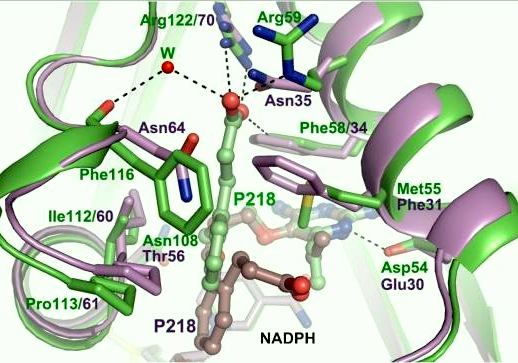
| - | There are three regions of the DHFR active site around the conserved arginine that differ between <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Human_dhfr_and_folate/1'>human</scene> and ''Plasmodium falciparum'' that allow a drug to target the PfDHFR specifically while not harming that of humans. The Met55, Cys/Arg59, and Phe116 residues in the PfDHFR are replaced by Phe31, Gln35, and Asn64 at structurally equivalent positions in human DHFR which are crucial differentiations that need to be recognized by inhibitors. The importance in devising a compound, such as P218, that would interact with the region surrounding the conserved Arg122 residue mentioned above is that the compound would interact with the PfDHFR Arg122 residue region while leaving the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Hdhfr_arg70/1'>human Arg70</scene> region and thus human DHFR alone. This design and intent for P218 was shown to be successful. In the quadruple mutant of PfDHFR, the P218 carboxylate side chain makes two hydrogen bonds with the Arg122 residue | + | There are three regions of the DHFR active site around the conserved arginine that differ between <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Human_dhfr_and_folate/1'>human</scene> and ''Plasmodium falciparum'' that allow a drug to target the PfDHFR specifically while not harming that of humans. The Met55, Cys/Arg59, and Phe116 residues in the PfDHFR are replaced by Phe31, Gln35, and Asn64 at structurally equivalent positions in human DHFR which are crucial differentiations that need to be recognized by inhibitors. The importance in devising a compound, such as P218, that would interact with the region surrounding the conserved Arg122 residue mentioned above is that the compound would interact with the PfDHFR Arg122 residue region while leaving the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Hdhfr_arg70/1'>human Arg70</scene> region and thus human DHFR alone. This design and intent for P218 was shown to be successful. In the quadruple mutant of PfDHFR, the P218 carboxylate side chain makes two hydrogen bonds with the Arg122 residue while in human DHFR, it has no interaction with the Arg70 residue. |
Revision as of 19:11, 29 November 2012
Introduction
Dihydrofolate reductase (DHFR) plays an essential role in the formation of DNA and has shown to be a reliable and one of the best targets for antimalarial drugs.[1] The current antimalarial drugs that target the malarial DHFR include pyrimethamine and cycloguanil. However, the effectiveness of these drugs has decreased because of mutations in the enzyme that have led to drug resistance. Since these mutations are becoming much more prevalent in malaria cases, new research in drug development must now incorporate both the wild-type as well as the quadruple mutant DHFR from the Plasmodium falciparum malarial strain, the most common and lethal of the malaria species.[2]
| |||||||||||
References
- ↑ Goodsell, David. "Dihydrofolate Reductase." RCSB PDB-101. RCSB PDB, Oct. 2002. Web. <http://www.rcsb.org/pdb/101/motm.do?momID=34>.
- ↑ Somsak V, Uthaipibull C, Prommana P, Srichairatanakool S, Yuthavong Y, Kamchonwongpaisan S. Transgenic Plasmodium parasites stably expressing Plasmodium vivax dihydrofolate reductase-thymidylate synthase as in vitro and in vivo models for antifolate screening. Malar J. 2011 Oct 7;10:291. PMID: 21981896
- ↑ Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012 Jul 28;11:243. PMID: 22839209
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
- ↑ Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. Epub 2012 Oct 3. PMID:23035243. doi: 10.1073/pnas.1204556109.
Proteopedia Page Contributors and Editors (what is this?)
Mary Smith, Alexander Berchansky, Karsten Theis, Michal Harel