Circadian Clock Protein KaiC
From Proteopedia
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
[[Image:KaiCABinteraction.jpg | thumb | alt=text | Interactions of KaiC with KaiA and KaiB ]] | [[Image:KaiCABinteraction.jpg | thumb | alt=text | Interactions of KaiC with KaiA and KaiB ]] | ||
| - | Biological Circadian Clocks are self-sustaining oscillators that function on a rhythmic cycle of or around 24 hours. | + | Biological Circadian Clocks are self-sustaining oscillators that function on a rhythmic cycle of or around 24 hours. They are found in almost all organisms, the simplest of which are cyanobacteria, which have been extensively studied in order to determine the mechanism of the fine-tuned biological process of circadian rhythmicity. In most eukaryotes, a region of the brain called the superchiasmic nuclei detects light and dark cycles, then relays the message to biological clock systems that maintain rhythmicity within the body. Conversely, cyanobacteria have a fairly modest system comprised of three proteins: KaiC, KaiA, and KaiB. The system is based around the central protein KaiC which exhibits ATP binding, inter-subunit organization, a scaffold region for Kai protein complex formation, a location where critical mutations are found, and an evolutionary link to other well-known proteins <ref name= Pattanayek>PMID: 15304218</ref>. In order to devise an explanation for the mechanism of biological oscillators, we need to characterize the structure, function, and interactions among molecular components. To study these, most scientists start with analyzing cyanobacteria, since it is the smallest organism that expresses circadian clock properties. |
==KaiC - KaiA - KaiB System== | ==KaiC - KaiA - KaiB System== | ||
| Line 13: | Line 13: | ||
==KaiC Autophosphorylation Sites== | ==KaiC Autophosphorylation Sites== | ||
| - | The phosphorylation sites on the KaiC protein are essential to the system. This is because phosphorylation status corresponds to clock speed. The protein predominantly phosphorylates on threonine and serine residues, whose specific identification is not completely resolved. Nonetheless, three potential phosphorylation sites have been identified within 10 Angstroms of the ATP binding region in the CII domain <ref name= Yao>PMCID: PMC518856</ref>. The key autophosphorylation site is <scene name='Circadian_Clock_Protein_KaiC/Active_site_thr432/1'>T432</scene>. The process is believed to demonstrate a transfer of the gamma phosphate of ATP from one CII subunit to the T432 site on an adjacent subunit. | + | The phosphorylation sites on the KaiC protein are essential to the system. This is because phosphorylation status corresponds to clock speed. The protein predominantly phosphorylates on threonine and serine residues, whose specific identification is not completely resolved. Nonetheless, three potential phosphorylation sites have been identified within 10 Angstroms of the ATP binding region in the CII domain <ref name= Yao>PMCID: PMC518856</ref>. The key autophosphorylation site is <scene name='Circadian_Clock_Protein_KaiC/Active_site_thr432/1'>T432</scene>. When this residue is mutated, there is no circadian rhythm at all. The process is believed to demonstrate a transfer of the gamma phosphate of ATP from one CII subunit to the T432 site on an adjacent subunit. |
A region of each hexamer that is notable regarding the phosphorylation of KaiC is the P Loop. This zone is recognized as site for binding and hydrolysis of ATP. Along with the T432 site, evidence shows a shuttling of phosphates between residues <scene name='Circadian_Clock_Protein_KaiC/Active_site_t432_t426_s431/1'>S431 and T426</scene> of the P Loop. | A region of each hexamer that is notable regarding the phosphorylation of KaiC is the P Loop. This zone is recognized as site for binding and hydrolysis of ATP. Along with the T432 site, evidence shows a shuttling of phosphates between residues <scene name='Circadian_Clock_Protein_KaiC/Active_site_t432_t426_s431/1'>S431 and T426</scene> of the P Loop. | ||
(difference between ATP binding and phosphorylation residues) | (difference between ATP binding and phosphorylation residues) | ||
| Line 31: | Line 31: | ||
==Biological Importance== | ==Biological Importance== | ||
- Nearly all promoters in a cyanobacteria are under circadian control. [function is important to whole life cycle] (1) | - Nearly all promoters in a cyanobacteria are under circadian control. [function is important to whole life cycle] (1) | ||
| - | + | - Structure similarity with RecA and DnaB. RecA is a DNA recombinase and DnaB is a DNA helicase, so the observation that there is similarity between these molecules imply possible direct interactions with DNA. | |
==References== | ==References== | ||
{{Reflist}} | {{Reflist}} | ||
Revision as of 07:16, 4 December 2012
Contents |
Introduction
Biological Circadian Clocks are self-sustaining oscillators that function on a rhythmic cycle of or around 24 hours. They are found in almost all organisms, the simplest of which are cyanobacteria, which have been extensively studied in order to determine the mechanism of the fine-tuned biological process of circadian rhythmicity. In most eukaryotes, a region of the brain called the superchiasmic nuclei detects light and dark cycles, then relays the message to biological clock systems that maintain rhythmicity within the body. Conversely, cyanobacteria have a fairly modest system comprised of three proteins: KaiC, KaiA, and KaiB. The system is based around the central protein KaiC which exhibits ATP binding, inter-subunit organization, a scaffold region for Kai protein complex formation, a location where critical mutations are found, and an evolutionary link to other well-known proteins [1]. In order to devise an explanation for the mechanism of biological oscillators, we need to characterize the structure, function, and interactions among molecular components. To study these, most scientists start with analyzing cyanobacteria, since it is the smallest organism that expresses circadian clock properties.
KaiC - KaiA - KaiB System
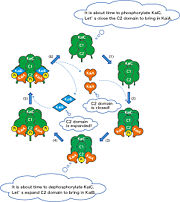
KaiC is the central clock protein, which has autokinase and autophosphorylase activity. Yet in the presence of ATP, KaiC cannot perform it's autophosphorylation function. It requires two other proteins, KaiA and KaiB, the genes of which are found in the same cluster on the chromosome [1]. Although KaiC phosphorylates itself, the presence of KaiA and KaiB are essential to rhythmicity. KaiA stimulates KaiC autophosphorylation, while KaiB antagonizes the process possibly by enhancing KaiC dephosphorylation. Even in the presence of high ATP, KaiB still prompts KaiC to dephosphorylate.
KaiC Homohexameric Complex
| |||||||||||
KaiA <-> KaiC Interaction Site
(show 3D image of KaiC site for KaiA binding and highlight key residues in interaction -weak? strong? any ions in site? how does it stabilize autophosporylation?) KaiA binds to the interface of the two donut-shaped KaiC subunits, CI and CII. This area, known as the "waist" of the molecule
KaiB <-> KaiC Interaction Site
(show 3D image of KaiC site for KaiB binding and highlight key residues in interaction -weak? strong? ions in site? how does it stabilize dephosphorylation/destabilize phosphorylation/destabilize KaiA?)
Biological Importance
- Nearly all promoters in a cyanobacteria are under circadian control. [function is important to whole life cycle] (1) - Structure similarity with RecA and DnaB. RecA is a DNA recombinase and DnaB is a DNA helicase, so the observation that there is similarity between these molecules imply possible direct interactions with DNA.
References
Proteopedia Page Contributors and Editors (what is this?)
Ashley Beechan, Michal Harel, Alexander Berchansky, Jaime Prilusky