Molecular Playground/OmpG
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:intactModelLargeText.jpg|frame|Bacterial chemotaxis receptor]] | [[Image:intactModelLargeText.jpg|frame|Bacterial chemotaxis receptor]] | ||
| - | + | Protein biosensors serve as an analytical device combining a biological component with a physiochemical detector. Through protein engineering we are able to utilize pores to be used as stochastic sensors, for single molecule detection (1). The applications of biosensors range from fundamental research, clinical diagnosis, and even advances in homeland security. PCR (Polymerase chain reaction) and ELISA (Enzyme- linked immunosorbent assay) are current sensitive detection methods. However, these methods are time consuming and require laborious effort, where results are provided hours or days later. For this reason alternate approaches are mounting in demands that are rapid in detection time, highly sensitive and reliable. | |
| + | Through stochastic sensing, utilizes the passage of ionic current through a protein pore containing engineered recognition sites, allowing for monitoring of analytes present (2). We propose the use of monomeric protein OmpG (outer membrane protein G) will allow us to tailor and fine tune properties of this pore in detection of analytes. This monomeric porin has features, which allow for complex properties to be customized as a sensor compartment, via protein engineering, providing eminent potential for the development of this protein as a successful biosensor. Furthermore, an OmpG sensing library could be built for the screening of constructs/ analytes according to their ability to recognize specific targets of medical relevance. | ||
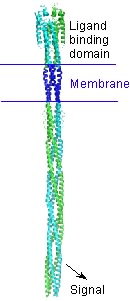
| - | A bacterial chemotaxis receptor is an unusually long alpha-helical structure. The attractant molecule (the ligand) binds near the top of this picture and sends a signal across the membrane into the cell to control proteins that bind near the bottom. This is a model of the structure of the receptor based on experimental structures of pieces of related proteins. | ||
Revision as of 19:41, 12 December 2012
Protein biosensors serve as an analytical device combining a biological component with a physiochemical detector. Through protein engineering we are able to utilize pores to be used as stochastic sensors, for single molecule detection (1). The applications of biosensors range from fundamental research, clinical diagnosis, and even advances in homeland security. PCR (Polymerase chain reaction) and ELISA (Enzyme- linked immunosorbent assay) are current sensitive detection methods. However, these methods are time consuming and require laborious effort, where results are provided hours or days later. For this reason alternate approaches are mounting in demands that are rapid in detection time, highly sensitive and reliable.
Through stochastic sensing, utilizes the passage of ionic current through a protein pore containing engineered recognition sites, allowing for monitoring of analytes present (2). We propose the use of monomeric protein OmpG (outer membrane protein G) will allow us to tailor and fine tune properties of this pore in detection of analytes. This monomeric porin has features, which allow for complex properties to be customized as a sensor compartment, via protein engineering, providing eminent potential for the development of this protein as a successful biosensor. Furthermore, an OmpG sensing library could be built for the screening of constructs/ analytes according to their ability to recognize specific targets of medical relevance.
|
Ligand-binding domain
The spinning protein () ) is the ligand binding domain of the aspartate receptor with the aspartate ligand bound (LKT).
Molecular Playground banner: A receptor protein used by bacteria to "smell" their environment.
| |||||||||||