Caspase-3 Regulatory Mechanisms
From Proteopedia
(→Caspase-3 Loop Bundle and Active Site) |
|||
| Line 18: | Line 18: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | == Caspase-3 Loop Bundle and Active Site== | ||
| + | |||
| + | <StructureSection load='2H5I' size='500' side='right' caption='Structure of Caspase-3 (PDB entry [[2H5I]])' scene='Caspase-3_Regulatory_Mechanisms/Scene1/1'> | ||
| + | === Importance of Loop Orientation=== | ||
| + | Caspases are extremely dependent on the orientation and geometry of their active site loops. If the loops are not ordered properly the enzyme fails to function. Caspase-3 has four active site loops on each half of the dimer constituting the active site bundle. Proteolytic activity is dependent on cleavage of an intersubunit linker, which releases loop 2 (L2) and L2’. <scene name='Caspase-3_Regulatory_Mechanisms/Scene2_nospin_labels/1'>L2'(green spheres) interacts with the opposite half of the dimer by holding up L2 (blue spheres) </scene>. This allows L2 to make critical contacts with L3 and L4, allowing them to organize the active site, bind substrate, and orient the nucleophilic cysteine 163 (bright green) so that it can cleave after aspartate residues. | ||
| + | |||
| + | |||
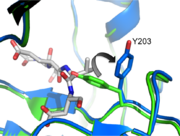
| + | [[Image:Y203flip.png | thumb| Tyrosine 203 Flip]] | ||
| + | |||
| + | Taking a closer look at L2 and L2’ we can see a critical interaction involving <scene name='Caspase-3_Regulatory_Mechanisms/D169_nospin/4'>Aspartate 169</scene> on L2. This residue makes two hydrogen bonds with backbone amides of V189’ and E190’, stabilizing L2 in the proper position. This reinforcement allows L2 to contact L3 so as to twist the active site cysteine into the proper orientation to attack the substrate. It also causes a conformational change at Tyrosine 203 (Tyrosine 203 Flip). The hydroxyl group occupies the P1 position in the active site, blocking substrate binding (green, pdb 1QX3). However, when L2 contacts L2' and finds the proper orientation, Y203 rotates 90 degrees and leaves a hole for P1 of the substrate (blue, pdb 2H5I). In addition, L2 can now contact L4 at K260. This secures L4 and allows it to make contacts in the P4 position, which greatly influence substrate specificity. | ||
| + | |||
| + | |||
| + | === Caspase-3 Active Site=== | ||
| + | |||
| + | The active site of caspase-3 utilizes a cysteine-histidine dyad, which has an exquisite specificity for cleaving after aspartate residues. Therefore, caspase-3, by definition, will have an aspartate in the <scene name='Caspase-3_Regulatory_Mechanisms/P1/2'>P1</scene> pocket. Uncleavable peptide substrates are often used in crystallography to bind to the active site. This will orient the delicate but deadly active site loops in order to facilitate the visualization of the chemistry of cleavage. The nucleophilic Cysteine 163 will work in concert with the second active site residue, Histidine 121, to attack the substrate. This reaction will ultimately cleave the peptide bond following the aspartate. | ||
| + | |||
| + | In order to be active and cleave the specific apoptotic targets, Caspase-3 must be able to first bind substrate. There are several essential interactions responsible for securing the substrate before cleavage. The binding pocket at <scene name='Caspase-3_Regulatory_Mechanisms/P2/2'>P2</scene> is a hydrophobic patch made up of Y204, W206, and F250 (dark blue residues). This creates a hydrophobic pocket for the P2 residue (in this casse, valine). At <scene name='Caspase-3_Regulatory_Mechanisms/P4/2'>P4</scene> there are contacts that contribute to the specificity of caspase-3. Asparagine 208 hydrogen bonds with an aspartate at P4 along with the backbone nitrogen of F250, creating a preference for a carboxylic acid at the P4 site. | ||
| + | |||
| + | |||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | |||
== Caspase-3 Regulation== | == Caspase-3 Regulation== | ||
| Line 48: | Line 72: | ||
Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466. | Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466. | ||
| - | |||
| - | </StructureSection> | ||
| - | |||
| - | |||
| - | == Caspase-3 Loop Bundle and Active Site== | ||
| - | |||
| - | <StructureSection load='2H5I' size='500' side='right' caption='Structure of Caspase-3 (PDB entry [[2H5I]])' scene='Caspase-3_Regulatory_Mechanisms/Scene1/1'> | ||
| - | === Importance of Loop Orientation=== | ||
| - | Caspases are extremely dependent on the orientation and geometry of their active site loops. If the loops are not ordered properly the enzyme fails to function. Caspase-3 has four active site loops on each half of the dimer constituting the active site bundle. Proteolytic activity is dependent on cleavage of an intersubunit linker, which releases loop 2 (L2) and L2’. <scene name='Caspase-3_Regulatory_Mechanisms/Scene2_nospin_labels/1'>L2'(green spheres) interacts with the opposite half of the dimer by holding up L2 (blue spheres) </scene>. This allows L2 to make critical contacts with L3 and L4, allowing them to organize the active site, bind substrate, and orient the nucleophilic cysteine 163 (bright green) so that it can cleave after aspartate residues. | ||
| - | |||
| - | |||
| - | [[Image:Y203flip.png | thumb| Tyrosine 203 Flip]] | ||
| - | |||
| - | Taking a closer look at L2 and L2’ we can see a critical interaction involving <scene name='Caspase-3_Regulatory_Mechanisms/D169_nospin/4'>Aspartate 169</scene> on L2. This residue makes two hydrogen bonds with backbone amides of V189’ and E190’, stabilizing L2 in the proper position. This reinforcement allows L2 to contact L3 so as to twist the active site cysteine into the proper orientation to attack the substrate. It also causes a conformational change at Tyrosine 203 (Tyrosine 203 Flip). The hydroxyl group occupies the P1 position in the active site, blocking substrate binding (green, pdb 1QX3). However, when L2 contacts L2' and finds the proper orientation, Y203 rotates 90 degrees and leaves a hole for P1 of the substrate (blue, pdb 2H5I). In addition, L2 can now contact L4 at K260. This secures L4 and allows it to make contacts in the P4 position, which greatly influence substrate specificity. | ||
| - | |||
| - | |||
| - | === Caspase-3 Active Site=== | ||
| - | |||
| - | The active site of caspase-3 utilizes a cysteine-histidine dyad, which has an exquisite specificity for cleaving after aspartate residues. Therefore, caspase-3, by definition, will have an aspartate in the <scene name='Caspase-3_Regulatory_Mechanisms/P1/2'>P1</scene> pocket. Uncleavable peptide substrates are often used in crystallography to bind to the active site. This will orient the delicate but deadly active site loops in order to facilitate the visualization of the chemistry of cleavage. The nucleophilic Cysteine 163 will work in concert with the second active site residue, Histidine 121, to attack the substrate. This reaction will ultimately cleave the peptide bond following the aspartate. | ||
| - | |||
| - | In order to be active and cleave the specific apoptotic targets, Caspase-3 must be able to first bind substrate. There are several essential interactions responsible for securing the substrate before cleavage. The binding pocket at <scene name='Caspase-3_Regulatory_Mechanisms/P2/2'>P2</scene> is a hydrophobic patch made up of Y204, W206, and F250 (dark blue residues). This creates a hydrophobic pocket for the P2 residue (in this casse, valine). At <scene name='Caspase-3_Regulatory_Mechanisms/P4/2'>P4</scene> there are contacts that contribute to the specificity of caspase-3. Asparagine 208 hydrogen bonds with an aspartate at P4 along with the backbone nitrogen of F250, creating a preference for a carboxylic acid at the P4 site. | ||
| - | |||
| - | |||
</StructureSection> | </StructureSection> | ||
Revision as of 00:36, 13 December 2012
Introduction
Caspases are cysteine-aspartic acid proteases and are key protein facilitators for the faithful execution of apoptosis or programmed cell death. Dysregulation in the apoptotic pathway has been implicated in a variety of diseases such as neurodegeneration, cancer, heart disease and some metabolic disorders. Because of the crucial role of caspases in the the apoptotic pathway, abnormalities in their functions would cause a haywire in the apoptotic cascade and can be deleterious to the cell. Caspases are thus being considered as therapeutic targets in apoptosis-related diseases.
Any apoptotic signal received by the cell causes the activation of initiator caspases (-8 and -9) by associating with another protein platform to form a functional holoenzyme. These initiator caspases then cleaves the executioner caspases -3, -6, -7. Caspase-3 specifically functions to cleave both caspase-6 and -7, which in turn cleave their respective targets to induce cell death. Aside from being able to activate caspase-6 and -7, caspase-3 also regulates caspase-9 activity, operating via feedback loop. These dual action of caspase-3 confers its distinct regulatory mechanisms, resulting a wider extent of effects in the apoptotic cascade.
| |||||||||||
Caspase-3 Loop Bundle and Active Site
| |||||||||||
Caspase-3 Regulation
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Scott Eron, Banyuhay P. Serrano, Alexander Berchansky, Yunlong Zhao, Jaime Prilusky, Michal Harel