Caspase-3 Regulatory Mechanisms
From Proteopedia
| Line 44: | Line 44: | ||
== Caspase-3 Regulation== | == Caspase-3 Regulation== | ||
| - | |||
<StructureSection load='1dq8' size='350' side='right' caption='Structure of Caspase-3 with substrate bound (PDB entry [[2H5I]])' scene='Caspase-3_Regulatory_Mechanisms/Scene1/1'> | <StructureSection load='1dq8' size='350' side='right' caption='Structure of Caspase-3 with substrate bound (PDB entry [[2H5I]])' scene='Caspase-3_Regulatory_Mechanisms/Scene1/1'> | ||
| - | |||
| - | == Regulation of Caspase-3== | ||
=== Exosite and Allosteric Site=== | === Exosite and Allosteric Site=== | ||
| Line 58: | Line 55: | ||
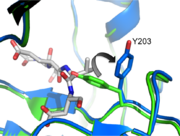
One interesting mutation, V266E, improves caspase-3 activity dramatically. Even in the uncleavable procaspase-3 (D5A, D26A, D175A), V266E mutant zymogen is still pseudo-activated, showing a 60-fold increase in activity. Intriguingly, V266E does not undergo a lot of conformational changes around the active site in its cleaved form. Based on the crystal structure of this mutant, the L2’ loop is partially disordered at 185’-180’. Moreover, this active procaspase-3 variant cannot be inhibited by endogenous XIAP like the normal cleaved caspase-3. So it provides us an option for apoptosis stimuli with intrinsic efficiency. | One interesting mutation, V266E, improves caspase-3 activity dramatically. Even in the uncleavable procaspase-3 (D5A, D26A, D175A), V266E mutant zymogen is still pseudo-activated, showing a 60-fold increase in activity. Intriguingly, V266E does not undergo a lot of conformational changes around the active site in its cleaved form. Based on the crystal structure of this mutant, the L2’ loop is partially disordered at 185’-180’. Moreover, this active procaspase-3 variant cannot be inhibited by endogenous XIAP like the normal cleaved caspase-3. So it provides us an option for apoptosis stimuli with intrinsic efficiency. | ||
| - | It was found recently that changing other critical residues (V266H, Y197C, E124A) on the dimer interface might play an important role on inhibition of caspase-3 by altering hydrogen bond patterns or through a relay of subtle conformational changes that | + | It was found recently that changing other critical residues (V266H, Y197C, E124A) on the dimer interface might play an important role on inhibition of caspase-3 by altering hydrogen bond patterns or through a relay of subtle conformational changes that spreads across the whole dimer. |
| + | |||
| + | === Post translational Modification=== | ||
| - | + | Caspase-3 has been reported to be phosphorylated at Ser-150 by p38-MAPK. | |
==== Natural Inhibitors==== | ==== Natural Inhibitors==== | ||
Revision as of 01:22, 13 December 2012
Contents |
Introduction
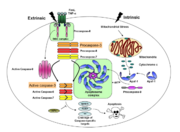
Caspases are cysteine-aspartic acid proteases and are key protein facilitators for the faithful execution of apoptosis or programmed cell death. Dysregulation in the apoptotic pathway has been implicated in a variety of diseases such as neurodegeneration, cancer, heart disease and some metabolic disorders. Because of the crucial role of caspases in the the apoptotic pathway, abnormalities in their functions would cause a haywire in the apoptotic cascade and can be deleterious to the cell. Caspases are thus being considered as therapeutic targets in apoptosis-related diseases.
Any apoptotic signal received by the cell causes the activation of initiator caspases (-8 and -9) by associating with other protein platforms to form a functional holoenzyme. These initiator caspases then cleaves the executioner caspases -3, -6, -7. Caspase-3 specifically functions to cleave both caspase-6 and -7, which in turn cleave their respective targets to induce cell death. Aside from being able to activate caspase-6 and -7, caspase-3 also regulates caspase-9 activity, operating via a feedback loop. These dual action of caspase-3 confers its distinct regulatory mechanisms, resulting a wider extent of its effects in the apoptotic cascade.
Overview of Caspase-3 Structure
| |||||||||||
Caspase-3 Loop Bundle and Active Site
| |||||||||||
Caspase-3 Regulation
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Scott Eron, Banyuhay P. Serrano, Alexander Berchansky, Yunlong Zhao, Jaime Prilusky, Michal Harel